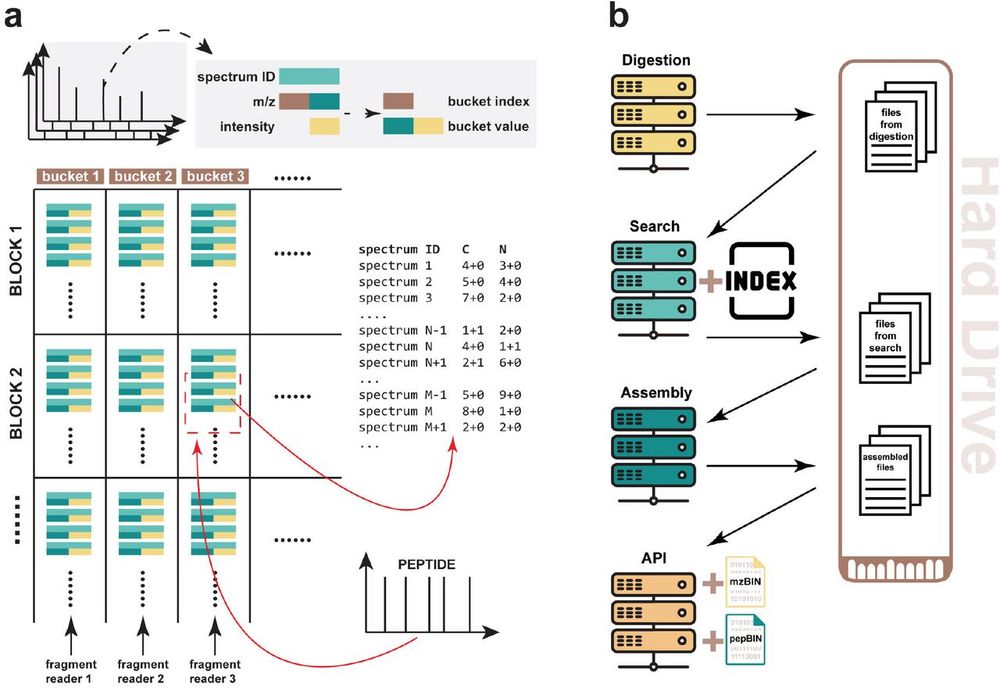
Our insights based on the data: www.biorxiv.org/content/10.6...
PRIDE repo will be made public in the next days.

Our insights based on the data: www.biorxiv.org/content/10.6...
PRIDE repo will be made public in the next days.
Release notes: github.com/vdemichev/Di...

Release notes: github.com/vdemichev/Di...
---
#proteomics #prot-paper

---
#proteomics #prot-paper
This serves as a basis for predicted spectral libraries, reducing the search space 10x-20x. www.biorxiv.org/content/10.1...

This serves as a basis for predicted spectral libraries, reducing the search space 10x-20x. www.biorxiv.org/content/10.1...
Learn more: bit.ly/4oz7Oaz

Learn more: bit.ly/4oz7Oaz

#PRIDE team is looking for a developer to build its future infrastructure. Our focus in a nutshell: #bigdata, single cell proteomics #SCP, & an #AI-oriented platform. Join us to shape the future of open proteomics data!
embl.wd103.myworkdayjobs.com/EMBL/job/Hin...

#PRIDE team is looking for a developer to build its future infrastructure. Our focus in a nutshell: #bigdata, single cell proteomics #SCP, & an #AI-oriented platform. Join us to shape the future of open proteomics data!
embl.wd103.myworkdayjobs.com/EMBL/job/Hin...


www.linkedin.com/posts/vadim-...

www.linkedin.com/posts/vadim-...
- A new contributor, Chris Bielow
- MaxDIA support, thanks @maxquant.bsky.social pmultiqc.quantms.org/MaxDIA/multi...
- DIANN reports way deeper thanks @vadim-demichev.bsky.social pmultiqc.quantms.org/DIANN/multiq...

- A new contributor, Chris Bielow
- MaxDIA support, thanks @maxquant.bsky.social pmultiqc.quantms.org/MaxDIA/multi...
- DIANN reports way deeper thanks @vadim-demichev.bsky.social pmultiqc.quantms.org/DIANN/multiq...

www.linkedin.com/pulse/pre-tr...

www.linkedin.com/pulse/pre-tr...

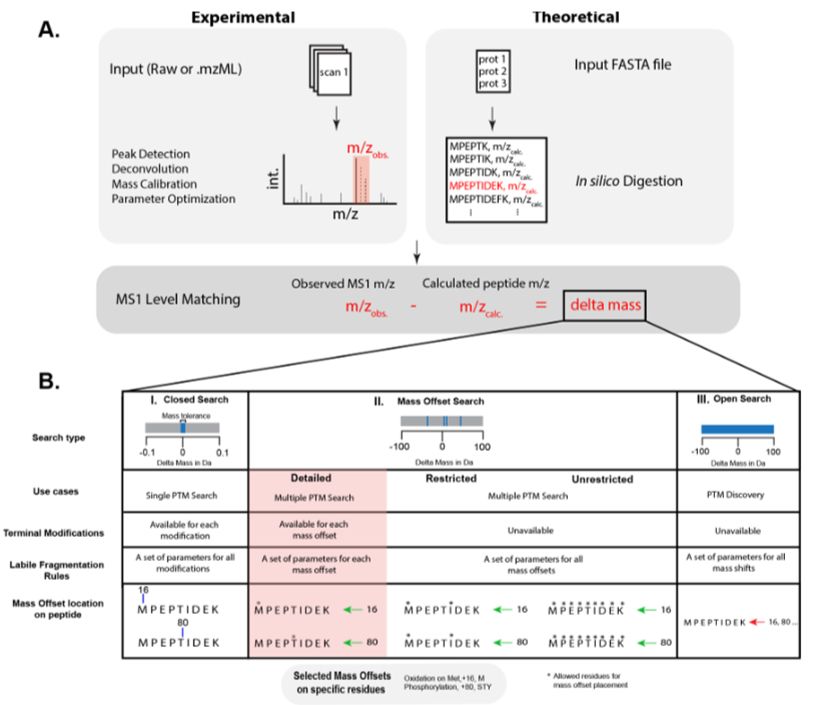

Presenting diaPASEF immunopeptidomics for bacterial epitope discovery.
💥 Showcasing DIA-NN immunopeptidomics using proteome-wide predicted HLA class I libraries!
🧬 Read more: biorxiv.org/cgi/content/...
#Immunopeptidomics #MassSpec #DIA #HLA

Presenting diaPASEF immunopeptidomics for bacterial epitope discovery.
💥 Showcasing DIA-NN immunopeptidomics using proteome-wide predicted HLA class I libraries!
🧬 Read more: biorxiv.org/cgi/content/...
#Immunopeptidomics #MassSpec #DIA #HLA
Patrick Willems @impenslab.bsky.social. With the new proteoform confidence module in DIA-NN, we enabled non-specific digest searches. Here's how it now works for immunopeptidomics!

Patrick Willems @impenslab.bsky.social. With the new proteoform confidence module in DIA-NN, we enabled non-specific digest searches. Here's how it now works for immunopeptidomics!

It is a minor update, main highlights are:
- Built-in support for Thermo .raw (Windows + Linux)
- New QC metrics in PDF report & visualising trends across the experiment

It is a minor update, main highlights are:
- Built-in support for Thermo .raw (Windows + Linux)
- New QC metrics in PDF report & visualising trends across the experiment