
Kusterlab
@kusterlab.bsky.social
Reposted by Kusterlab
Exited to share our latest work! Out now in @natcomms.nature.com
Koina aims to transform how #proteomics uses machine learning. You no longer need to be a tech wizard to use ML and now can easily run #ML models. Integrated with FragPipe, Skyline and EncyclopeDIA!
www.nature.com/articles/s41...
Koina aims to transform how #proteomics uses machine learning. You no longer need to be a tech wizard to use ML and now can easily run #ML models. Integrated with FragPipe, Skyline and EncyclopeDIA!
www.nature.com/articles/s41...

Koina: Democratizing machine learning for proteomics research - Nature Communications
Koina is an open-source, online platform that simplifies access to machine learning models in proteomics, enabling easier integration into analysis tools and helping researchers adopt and reuse ML mod...
www.nature.com
November 11, 2025 at 8:06 PM
Exited to share our latest work! Out now in @natcomms.nature.com
Koina aims to transform how #proteomics uses machine learning. You no longer need to be a tech wizard to use ML and now can easily run #ML models. Integrated with FragPipe, Skyline and EncyclopeDIA!
www.nature.com/articles/s41...
Koina aims to transform how #proteomics uses machine learning. You no longer need to be a tech wizard to use ML and now can easily run #ML models. Integrated with FragPipe, Skyline and EncyclopeDIA!
www.nature.com/articles/s41...
Hello Toronto!🍁The Terrific TUM Team is happy to be attending #HUPO2025. Kusterlab, Wilhelmlab and Leelab have a diverse set of presentations for you that we can't wait to share 🤓.
We're excited to spend the next days reuniting with old friends and making new connections. See you there!
We're excited to spend the next days reuniting with old friends and making new connections. See you there!

November 9, 2025 at 11:33 AM
Hello Toronto!🍁The Terrific TUM Team is happy to be attending #HUPO2025. Kusterlab, Wilhelmlab and Leelab have a diverse set of presentations for you that we can't wait to share 🤓.
We're excited to spend the next days reuniting with old friends and making new connections. See you there!
We're excited to spend the next days reuniting with old friends and making new connections. See you there!
Our new review paper about Pathway-Centric PTM Data Analysis is out this week in #Proteomics!
We cover databases, enrichment tools, software for pathway reconstruction, and full-fledged platforms that help to interpret high-throughput PTM datasets.
Check it out here: doi.org/10.1002/pmic...
We cover databases, enrichment tools, software for pathway reconstruction, and full-fledged platforms that help to interpret high-throughput PTM datasets.
Check it out here: doi.org/10.1002/pmic...

Computational Approaches for Pathway‐Centric Analysis of Protein Post‐Translational Modifications
Protein function is dynamically modulated by post-translational modifications (PTMs). Many different types of PTMs can nowadays be identified and quantified at a large scale using mass spectrometry. ...
doi.org
October 20, 2025 at 7:07 AM
Our new review paper about Pathway-Centric PTM Data Analysis is out this week in #Proteomics!
We cover databases, enrichment tools, software for pathway reconstruction, and full-fledged platforms that help to interpret high-throughput PTM datasets.
Check it out here: doi.org/10.1002/pmic...
We cover databases, enrichment tools, software for pathway reconstruction, and full-fledged platforms that help to interpret high-throughput PTM datasets.
Check it out here: doi.org/10.1002/pmic...
New preprint: We isolate peptide–RNA photo-crosslinks with tunable RNA chains from living cells for mass spec. This maps over 4,700 crosslinking sites across 744 proteins and offers the first glimpse into the RNA sequences in crosslinks by MS. Read here: doi.org/10.1101/2025...

Peptide-RNA photo-crosslinks with tunable RNA chain map protein-RNA interfaces
Photo-crosslinking mass spectrometry enables the identification of protein-RNA interactions in living cells, pinpointing interaction interfaces at single-amino acid resolution. However, current isolat...
doi.org
September 9, 2025 at 11:46 AM
New preprint: We isolate peptide–RNA photo-crosslinks with tunable RNA chains from living cells for mass spec. This maps over 4,700 crosslinking sites across 744 proteins and offers the first glimpse into the RNA sequences in crosslinks by MS. Read here: doi.org/10.1101/2025...
New Preprint: High-intensity 365 nm irradiation accelerates photoreactions in living cells by up to 1000×. We show protein–drug 💊, –protein 💪, –DNA 🧬, –RNA 🧬 crosslinking within seconds, and analysis by #massspec for #proteomics, #chembio, #chromatin and #RNAbiology. www.biorxiv.org/content/10.1...

Enhanced photo-crosslinking in living cells with high-intensity longwave ultraviolet light
The activation of chemical reactions in living cells using ultraviolet (UV) light enables the interrogation of biomolecules in their native environment with photoreactive probes or crosslinking reagen...
www.biorxiv.org
August 25, 2025 at 12:07 PM
New Preprint: High-intensity 365 nm irradiation accelerates photoreactions in living cells by up to 1000×. We show protein–drug 💊, –protein 💪, –DNA 🧬, –RNA 🧬 crosslinking within seconds, and analysis by #massspec for #proteomics, #chembio, #chromatin and #RNAbiology. www.biorxiv.org/content/10.1...
Thanks @tum.de for highlighting Jakob's recent paper in @cellpress.bsky.social in the university news:
www.ls.tum.de/en/ls/public...
If you haven't had the chance to read the publication, you can check it out here: doi.org/10.1016/j.ce...
www.ls.tum.de/en/ls/public...
If you haven't had the chance to read the publication, you can check it out here: doi.org/10.1016/j.ce...

August 7, 2025 at 12:26 PM
Thanks @tum.de for highlighting Jakob's recent paper in @cellpress.bsky.social in the university news:
www.ls.tum.de/en/ls/public...
If you haven't had the chance to read the publication, you can check it out here: doi.org/10.1016/j.ce...
www.ls.tum.de/en/ls/public...
If you haven't had the chance to read the publication, you can check it out here: doi.org/10.1016/j.ce...
Excited to share our latest work published in #ScienceSignaling! 🚀
www.science.org/doi/10.1126/...
(1/4)
www.science.org/doi/10.1126/...
(1/4)
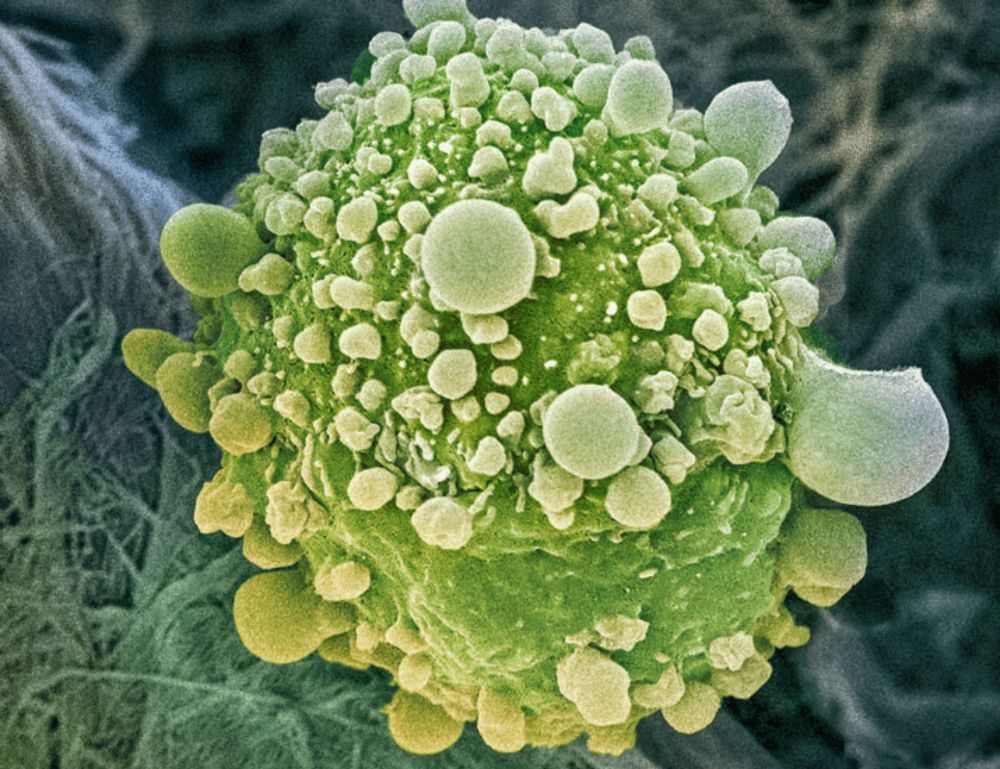
Proteomic analyses identify targets, pathways, and cellular consequences of oncogenic KRAS signaling
Proteomic analyses offer insights into how KRAS inhibitors affect oncogenic KRAS signaling.
www.science.org
July 31, 2025 at 8:10 AM
Excited to share our latest work published in #ScienceSignaling! 🚀
www.science.org/doi/10.1126/...
(1/4)
www.science.org/doi/10.1126/...
(1/4)
Kusterlab is looking for a new member to join our Bioinformatics team.
Apply now if you're interested in working with us!
Apply now if you're interested in working with us!
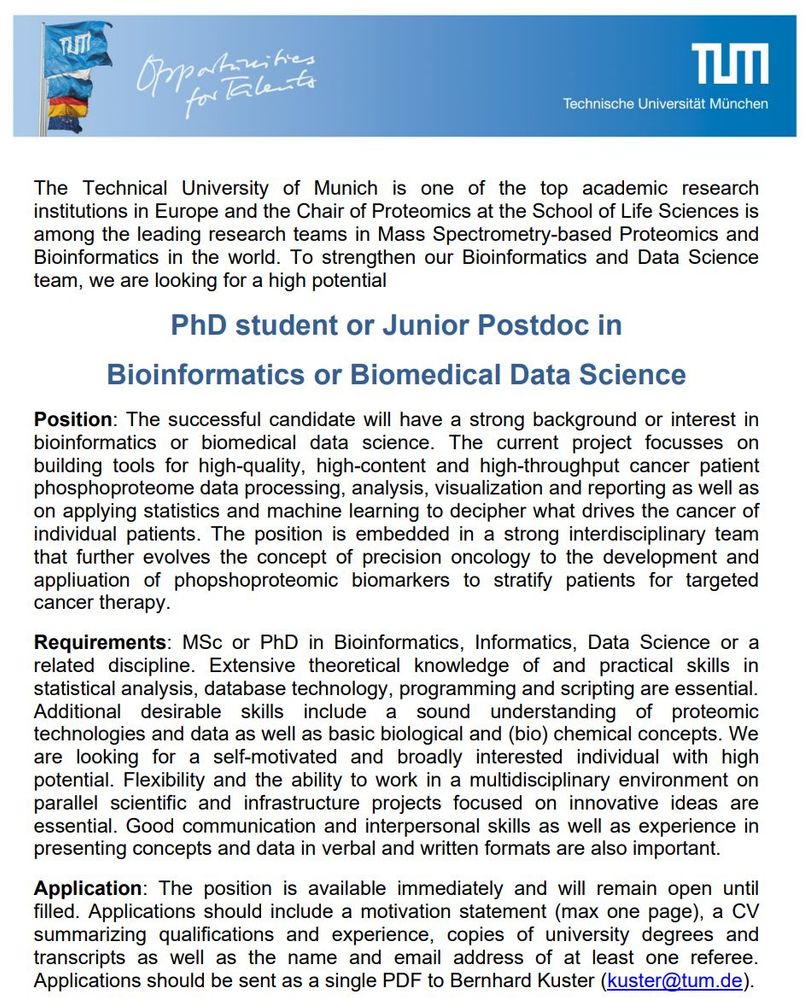
July 30, 2025 at 12:56 PM
Kusterlab is looking for a new member to join our Bioinformatics team.
Apply now if you're interested in working with us!
Apply now if you're interested in working with us!
Another day of #ASMS2025 in Baltimore, and we're back with another poster! Make sure to visit Flo today - he'll present the latest insights from his large-scale decryptM project.
Go #TeamMassSpec!
Go #TeamMassSpec!

June 4, 2025 at 9:28 AM
Another day of #ASMS2025 in Baltimore, and we're back with another poster! Make sure to visit Flo today - he'll present the latest insights from his large-scale decryptM project.
Go #TeamMassSpec!
Go #TeamMassSpec!
Hello Baltimore! We're looking forward to #ASMS2025 and all the new developments in mass-spec and proteomics.
If you're attending today, make sure to check out @msleelab.bsky.social's poster on citrullination in multiple sclerosis. Sophia 'Lapo' Laposchan is excited to see you there!
If you're attending today, make sure to check out @msleelab.bsky.social's poster on citrullination in multiple sclerosis. Sophia 'Lapo' Laposchan is excited to see you there!

June 2, 2025 at 8:14 AM
Hello Baltimore! We're looking forward to #ASMS2025 and all the new developments in mass-spec and proteomics.
If you're attending today, make sure to check out @msleelab.bsky.social's poster on citrullination in multiple sclerosis. Sophia 'Lapo' Laposchan is excited to see you there!
If you're attending today, make sure to check out @msleelab.bsky.social's poster on citrullination in multiple sclerosis. Sophia 'Lapo' Laposchan is excited to see you there!
🚨Our new paper is online🚨
We use zero-distance⚡photo-crosslinking⚡to reveal direct protein-DNA interactions in living cells, enabling quantitative analysis of the DNA-interacting proteome on a timescale of minutes. #DNA #Chromatin #Proteomics
www.cell.com/cell/fulltex...
We use zero-distance⚡photo-crosslinking⚡to reveal direct protein-DNA interactions in living cells, enabling quantitative analysis of the DNA-interacting proteome on a timescale of minutes. #DNA #Chromatin #Proteomics
www.cell.com/cell/fulltex...
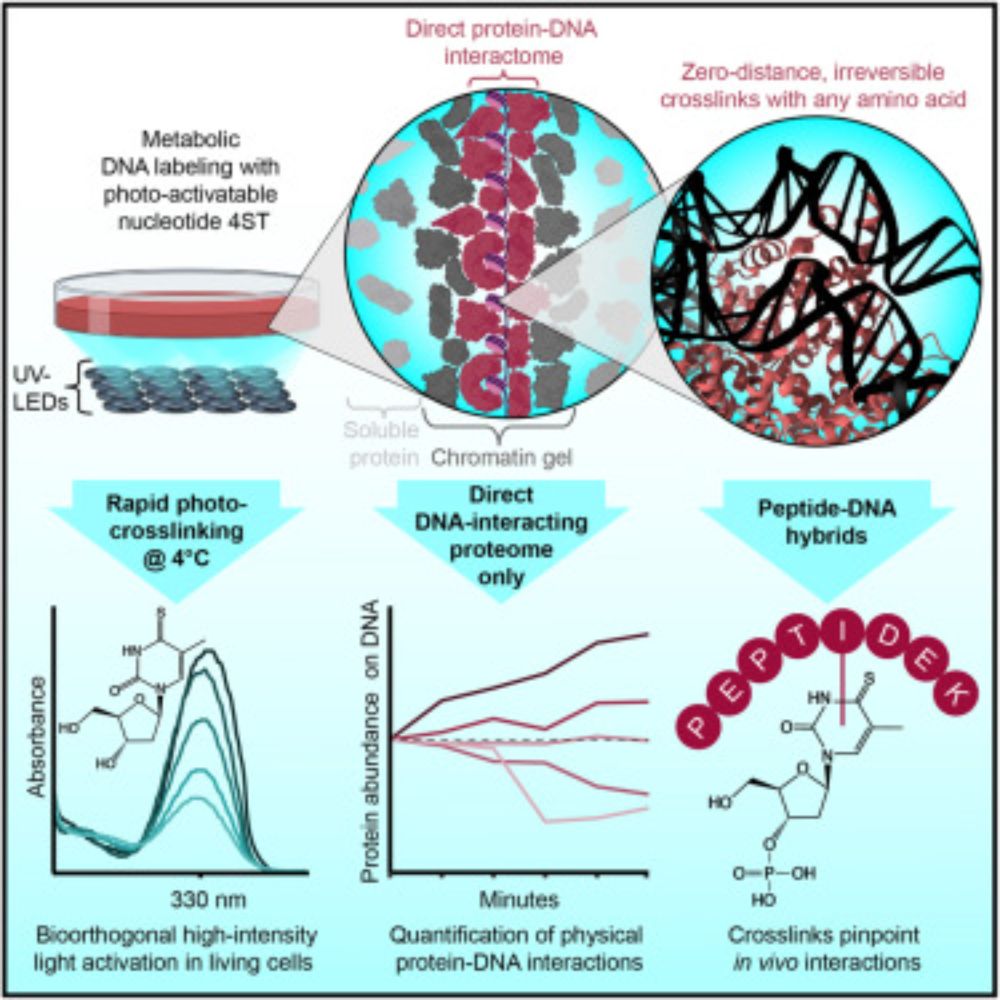
The human proteome with direct physical access to DNA
Zero-distance photo-crosslinking reveals direct protein-DNA interactions in living
cells, enabling quantitative analysis of the DNA-interacting proteome on a timescale
of minutes with single-amino-aci...
www.cell.com
May 22, 2025 at 6:44 PM
🚨Our new paper is online🚨
We use zero-distance⚡photo-crosslinking⚡to reveal direct protein-DNA interactions in living cells, enabling quantitative analysis of the DNA-interacting proteome on a timescale of minutes. #DNA #Chromatin #Proteomics
www.cell.com/cell/fulltex...
We use zero-distance⚡photo-crosslinking⚡to reveal direct protein-DNA interactions in living cells, enabling quantitative analysis of the DNA-interacting proteome on a timescale of minutes. #DNA #Chromatin #Proteomics
www.cell.com/cell/fulltex...
New paper!
Our chemoproteomic survey of phenylhydroxamic acids identifies the first drug-like inhibitors for nucleotide-binding protein HINT1 and nucleoside kinases NME1-4. On top, we found probes for the HDAC inhibitor off-target MBLAC2. Check out the open-access article! tinyurl.com/yh92vh6b
Our chemoproteomic survey of phenylhydroxamic acids identifies the first drug-like inhibitors for nucleotide-binding protein HINT1 and nucleoside kinases NME1-4. On top, we found probes for the HDAC inhibitor off-target MBLAC2. Check out the open-access article! tinyurl.com/yh92vh6b
May 9, 2025 at 12:41 PM
New paper!
Our chemoproteomic survey of phenylhydroxamic acids identifies the first drug-like inhibitors for nucleotide-binding protein HINT1 and nucleoside kinases NME1-4. On top, we found probes for the HDAC inhibitor off-target MBLAC2. Check out the open-access article! tinyurl.com/yh92vh6b
Our chemoproteomic survey of phenylhydroxamic acids identifies the first drug-like inhibitors for nucleotide-binding protein HINT1 and nucleoside kinases NME1-4. On top, we found probes for the HDAC inhibitor off-target MBLAC2. Check out the open-access article! tinyurl.com/yh92vh6b
New paper!
Our chemoproteomic survey of phenylhydroxamic acids identifies the first drug-like inhibitors for nucleotide-binding protein HINT1 and nucleoside kinases NME1-4. On top, we found probes for the HDAC inhibitor off-target MBLAC2. Check out the open-access article! tinyurl.com/yh92vh6b
Our chemoproteomic survey of phenylhydroxamic acids identifies the first drug-like inhibitors for nucleotide-binding protein HINT1 and nucleoside kinases NME1-4. On top, we found probes for the HDAC inhibitor off-target MBLAC2. Check out the open-access article! tinyurl.com/yh92vh6b

sev on X: "Paper alert! We report the first inhibitors for nucleoside kinases NME1-4 and for the nucleotide-binding protein HINT1. On top, we provide probes for MBLAC2, an off-target of every second (!) hydroxamic acid-based HDAC inhibitor! https://t.co/uuRCO78xFz 👇🧵" / X
Paper alert! We report the first inhibitors for nucleoside kinases NME1-4 and for the nucleotide-binding protein HINT1. On top, we provide probes for MBLAC2, an off-target of every second (!) hydroxamic acid-based HDAC inhibitor! https://t.co/uuRCO78xFz 👇🧵
x.com
May 9, 2025 at 11:33 AM
New paper!
Our chemoproteomic survey of phenylhydroxamic acids identifies the first drug-like inhibitors for nucleotide-binding protein HINT1 and nucleoside kinases NME1-4. On top, we found probes for the HDAC inhibitor off-target MBLAC2. Check out the open-access article! tinyurl.com/yh92vh6b
Our chemoproteomic survey of phenylhydroxamic acids identifies the first drug-like inhibitors for nucleotide-binding protein HINT1 and nucleoside kinases NME1-4. On top, we found probes for the HDAC inhibitor off-target MBLAC2. Check out the open-access article! tinyurl.com/yh92vh6b
One algorithm to rule them all? CHIMERYS bridges the gap — DDA,DIA and PRM — together at last!
With #CHIMERYS, we can now directly compare DDA and DIA data — 🍎 to 🍎 finally made possible.
doi.org/10.1038/s41592-025-02663-w
#KusterLab #WilhelmLab #MSAID #Proteomics
With #CHIMERYS, we can now directly compare DDA and DIA data — 🍎 to 🍎 finally made possible.
doi.org/10.1038/s41592-025-02663-w
#KusterLab #WilhelmLab #MSAID #Proteomics
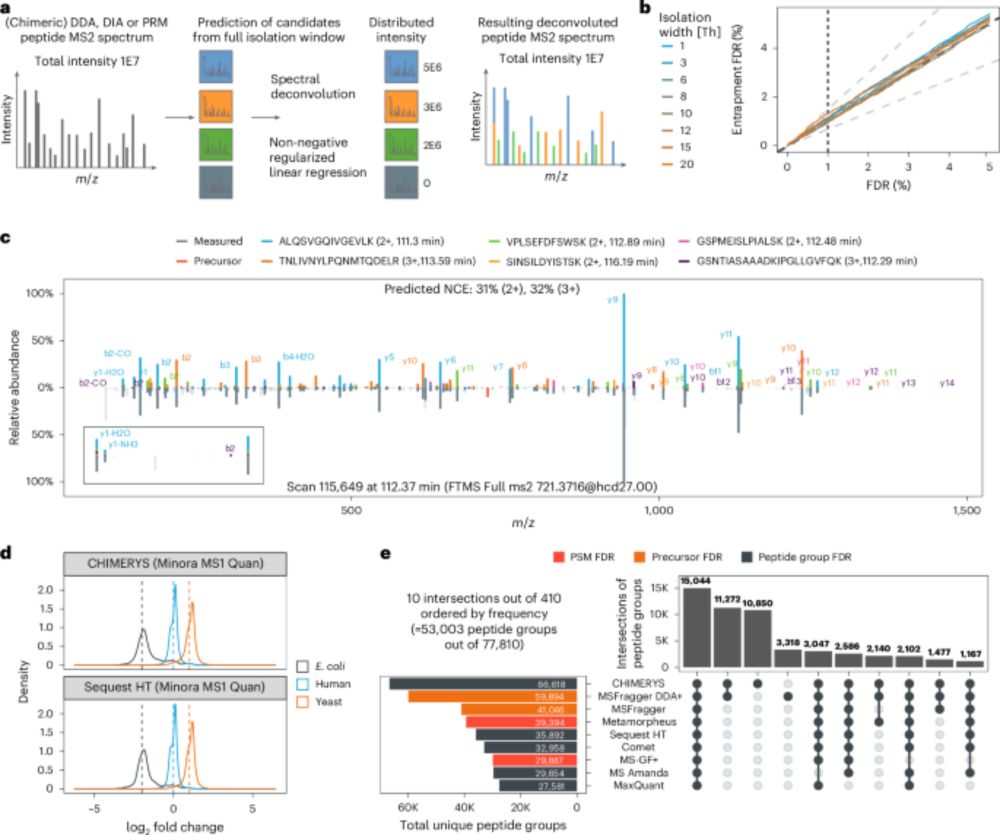
Unifying the analysis of bottom-up proteomics data with CHIMERYS - Nature Methods
CHIMERYS is a spectrum-centric and data acquisition method-agnostic algorithm for the analysis of MS2 spectra. It is capable of deconvoluting any MS2 spectrum, regardless of whether it was acquired by...
doi.org
April 22, 2025 at 11:44 AM
One algorithm to rule them all? CHIMERYS bridges the gap — DDA,DIA and PRM — together at last!
With #CHIMERYS, we can now directly compare DDA and DIA data — 🍎 to 🍎 finally made possible.
doi.org/10.1038/s41592-025-02663-w
#KusterLab #WilhelmLab #MSAID #Proteomics
With #CHIMERYS, we can now directly compare DDA and DIA data — 🍎 to 🍎 finally made possible.
doi.org/10.1038/s41592-025-02663-w
#KusterLab #WilhelmLab #MSAID #Proteomics
Reposted by Kusterlab
3/2025 Issue ➡️ www.embopress.org/toc/17444292...
metabolic mutations drive bacterial antibiotic resistance evolution, prior knowledge for GRN inference, HPA drugs fail mood disorder clinical trials
Cover: ATR kinase inhibitors work together to overcome chemoresistance @kusterlab.bsky.social
metabolic mutations drive bacterial antibiotic resistance evolution, prior knowledge for GRN inference, HPA drugs fail mood disorder clinical trials
Cover: ATR kinase inhibitors work together to overcome chemoresistance @kusterlab.bsky.social

March 12, 2025 at 9:56 AM
3/2025 Issue ➡️ www.embopress.org/toc/17444292...
metabolic mutations drive bacterial antibiotic resistance evolution, prior knowledge for GRN inference, HPA drugs fail mood disorder clinical trials
Cover: ATR kinase inhibitors work together to overcome chemoresistance @kusterlab.bsky.social
metabolic mutations drive bacterial antibiotic resistance evolution, prior knowledge for GRN inference, HPA drugs fail mood disorder clinical trials
Cover: ATR kinase inhibitors work together to overcome chemoresistance @kusterlab.bsky.social
🚀 Exciting news from our lab! Our latest paper has been featured on the cover of @molsystbiol.org - "Gemcitabine and ATR inhibitors synergize to kill PDAC cells by blocking DNA damage response" by Höfer et al.! 🧬🎉 doi.org/10.1038/s44320-025-00085-6 (1/4)

Gemcitabine and ATR inhibitors synergize to kill PDAC cells by blocking DNA damage response | Molecular Systems Biology
imageimagePhosphoproteomics unveils the mode of action of clinical ATR inhibitors and explains
their synergy with Gemcitabine in pancreatic cancer cells. Viability screening of 146 targeted drugs ide...
doi.org
March 4, 2025 at 4:06 PM
🚀 Exciting news from our lab! Our latest paper has been featured on the cover of @molsystbiol.org - "Gemcitabine and ATR inhibitors synergize to kill PDAC cells by blocking DNA damage response" by Höfer et al.! 🧬🎉 doi.org/10.1038/s44320-025-00085-6 (1/4)
🎉 We're happy to announce that our latest project was published in @naturecomms.bsky.social this week: PTMNavigator, a #bioinformatics web platform for in-depth analysis of post-translational modification (PTM) perturbation datasets.
📄 doi.org/10.1038/s414... (1/6)
📄 doi.org/10.1038/s414... (1/6)
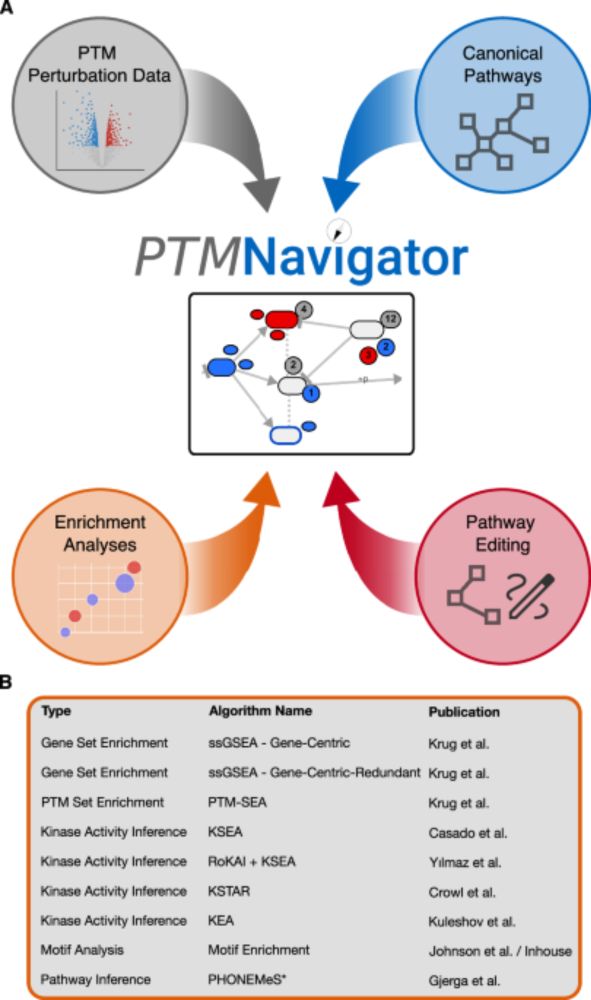
PTMNavigator: interactive visualization of differentially regulated post-translational modifications in cellular signaling pathways - Nature Communications
Post-translational modifications are important regulators of cellular pathways, but our understanding of these processes is limited. Here, the authors present a web tool that integrates various databa...
doi.org
January 10, 2025 at 2:20 PM
🎉 We're happy to announce that our latest project was published in @naturecomms.bsky.social this week: PTMNavigator, a #bioinformatics web platform for in-depth analysis of post-translational modification (PTM) perturbation datasets.
📄 doi.org/10.1038/s414... (1/6)
📄 doi.org/10.1038/s414... (1/6)
(1/5)
🚨 New Research Alert! 🚨
Excited to share our latest work in @embojournal.bsky.social
"Towards routine proteome profiling of FFPE tissue: insights from a 1,220-case pan-cancer study" ✨
doi.org/10.1038/s443...
📌 Key Highlights:
🚨 New Research Alert! 🚨
Excited to share our latest work in @embojournal.bsky.social
"Towards routine proteome profiling of FFPE tissue: insights from a 1,220-case pan-cancer study" ✨
doi.org/10.1038/s443...
📌 Key Highlights:

Towards routine proteome profiling of FFPE tissue: insights from a 1,220-case pan-cancer study | The EMBO Journal
imageimageFormalin fixed paraffin embedded (FFPE) patient tumor specimens are readily accessible
and established gold standard for cancer diagnostics in pathology laboratories. To
start building a pan...
doi.org
December 13, 2024 at 11:38 AM
(1/5)
🚨 New Research Alert! 🚨
Excited to share our latest work in @embojournal.bsky.social
"Towards routine proteome profiling of FFPE tissue: insights from a 1,220-case pan-cancer study" ✨
doi.org/10.1038/s443...
📌 Key Highlights:
🚨 New Research Alert! 🚨
Excited to share our latest work in @embojournal.bsky.social
"Towards routine proteome profiling of FFPE tissue: insights from a 1,220-case pan-cancer study" ✨
doi.org/10.1038/s443...
📌 Key Highlights:
That's a wrap on #HUPO2024! Kusterlab, Leelab, Wilhelmlab, and BayBioMS had an awesome time in Dresden. It was good to meet so many familiar and new faces in the community. See you next time! https://t.co/n306Mxe1fw
November 25, 2024 at 7:43 AM
That's a wrap on #HUPO2024! Kusterlab, Leelab, Wilhelmlab, and BayBioMS had an awesome time in Dresden. It was good to meet so many familiar and new faces in the community. See you next time! https://t.co/n306Mxe1fw
#HUPO2024 starts tomorrow and the Terrific TUM Team is ready! Kusterlab, Wilhelmlab (@wilhelm_compms), Leelab (@msleemslab) and BayBioMS (@BayBioMS) have printed their posters and prepared their talks, and we're very excited for the next days. See you in Dresden! https://t.co/RtEIuAOLok
November 25, 2024 at 7:43 AM
#HUPO2024 starts tomorrow and the Terrific TUM Team is ready! Kusterlab, Wilhelmlab (@wilhelm_compms), Leelab (@msleemslab) and BayBioMS (@BayBioMS) have printed their posters and prepared their talks, and we're very excited for the next days. See you in Dresden! https://t.co/RtEIuAOLok
Shoutout to our colleagues at @proteomicsdb who are working overtime to solve this!
November 25, 2024 at 7:43 AM
Shoutout to our colleagues at @proteomicsdb who are working overtime to solve this!
ProteomicsDB has had some downtime lately because of hardware issues. We're sorry for that and promise that it will be up and running again soon! https://t.co/kN4uJ2gMxA
November 25, 2024 at 7:43 AM
ProteomicsDB has had some downtime lately because of hardware issues. We're sorry for that and promise that it will be up and running again soon! https://t.co/kN4uJ2gMxA
... and it's back: https://t.co/1tFPnJWsyv
November 25, 2024 at 7:43 AM
... and it's back: https://t.co/1tFPnJWsyv
Our friends at https://t.co/1tFPnJWsyv are rolling out an update. Stay tuned for new content and features 👀 https://t.co/fpp2LUm3NI
November 25, 2024 at 7:43 AM
Our friends at https://t.co/1tFPnJWsyv are rolling out an update. Stay tuned for new content and features 👀 https://t.co/fpp2LUm3NI
Final Day of #ASMS2024 in Anaheim, but the Terrific TUM Team is far from done! Ludwig from Wilhelmlab will tell you all about Koina today. And at their posters, meet Flo (Kusterlab), Joel (Wilhelmlab), and Miri (@BayBioMS). See you there! One final time: Go #TeamMassSpec! https://t.co/yeX48Go2nZ
November 25, 2024 at 7:43 AM
Final Day of #ASMS2024 in Anaheim, but the Terrific TUM Team is far from done! Ludwig from Wilhelmlab will tell you all about Koina today. And at their posters, meet Flo (Kusterlab), Joel (Wilhelmlab), and Miri (@BayBioMS). See you there! One final time: Go #TeamMassSpec! https://t.co/yeX48Go2nZ

