We're excited to spend the next days reuniting with old friends and making new connections. See you there!

We're excited to spend the next days reuniting with old friends and making new connections. See you there!
www.ls.tum.de/en/ls/public...
If you haven't had the chance to read the publication, you can check it out here: doi.org/10.1016/j.ce...

www.ls.tum.de/en/ls/public...
If you haven't had the chance to read the publication, you can check it out here: doi.org/10.1016/j.ce...
Apply now if you're interested in working with us!
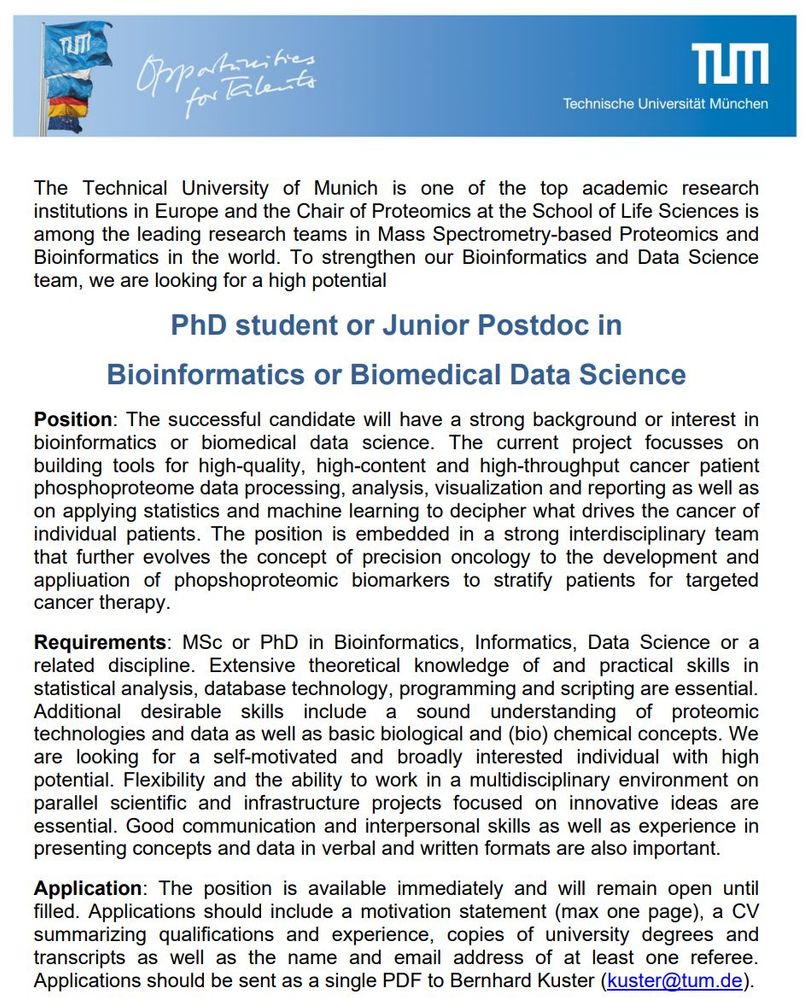
Apply now if you're interested in working with us!
Go #TeamMassSpec!

Go #TeamMassSpec!
If you're attending today, make sure to check out @msleelab.bsky.social's poster on citrullination in multiple sclerosis. Sophia 'Lapo' Laposchan is excited to see you there!

If you're attending today, make sure to check out @msleelab.bsky.social's poster on citrullination in multiple sclerosis. Sophia 'Lapo' Laposchan is excited to see you there!
Thanks to everyone involved! 🥳 We're looking forward to seeing how it'll be received by the community. (6/6).

Thanks to everyone involved! 🥳 We're looking forward to seeing how it'll be received by the community. (6/6).
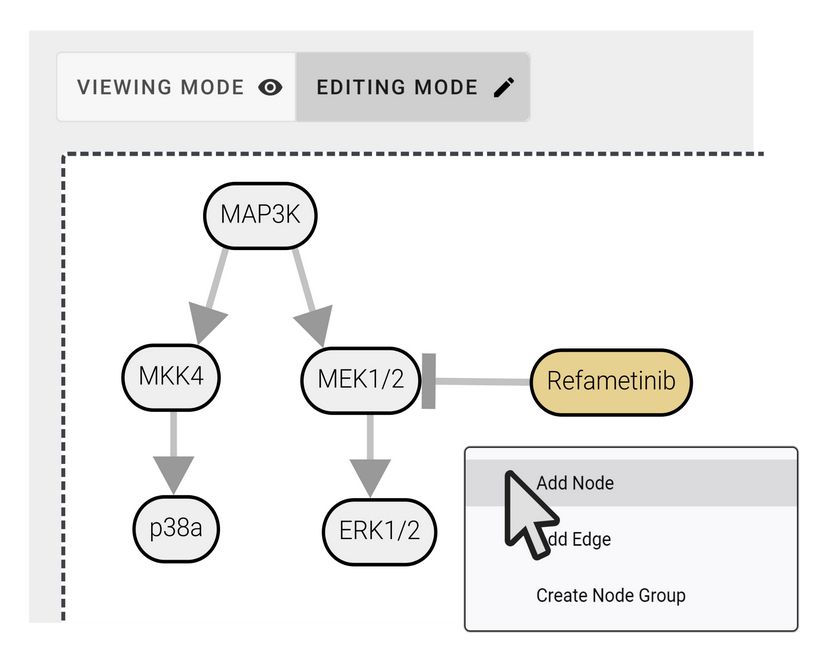
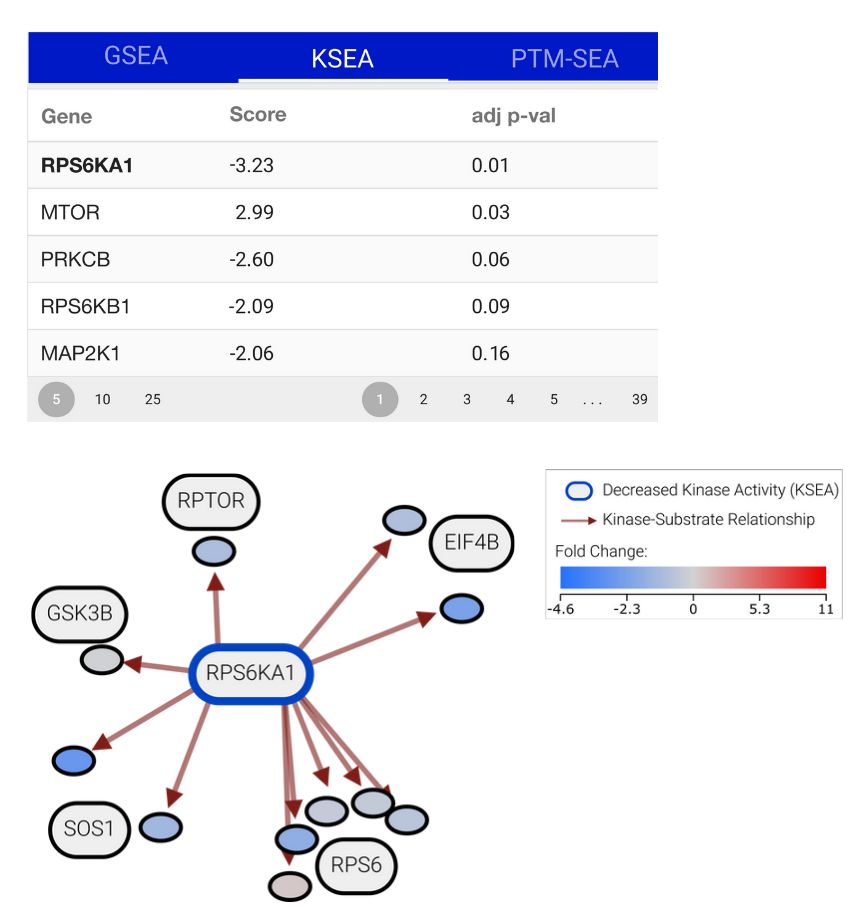
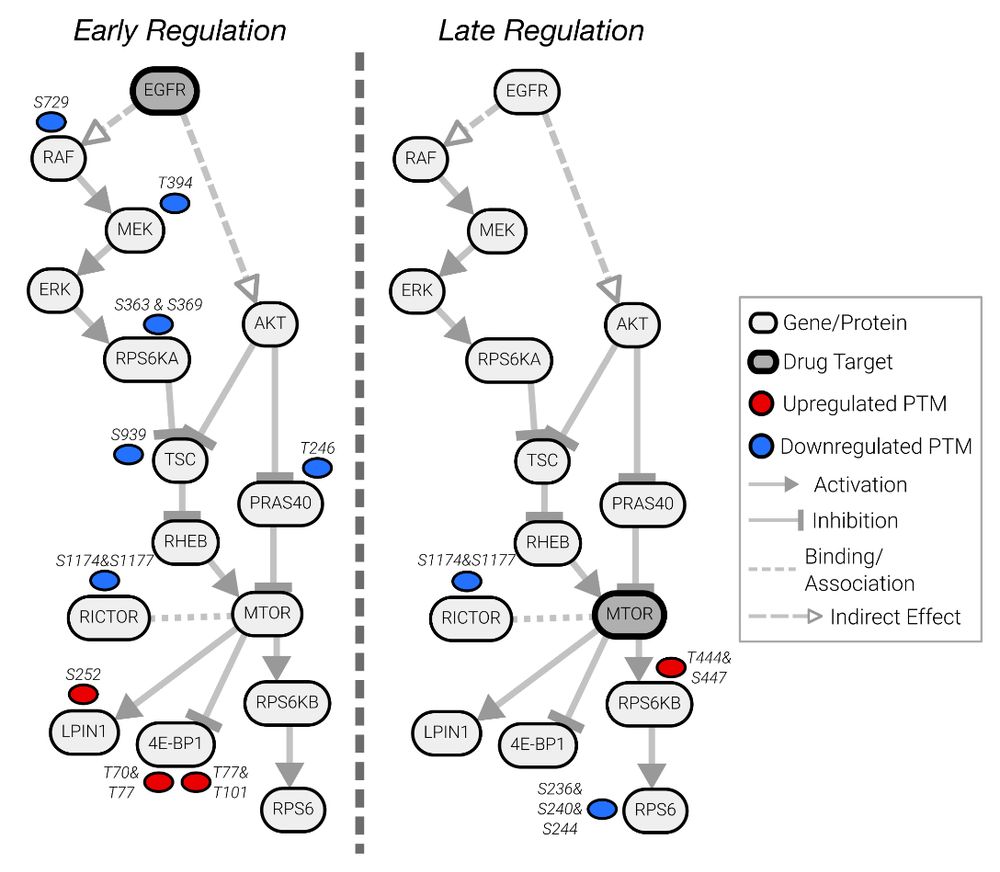
To tackle this, Julian Müller and colleagues have developed PTMNavigator, which is available at ProteomicsDB 💻 www.proteomicsdb.org/analytics/ptmNavigator (2/6)
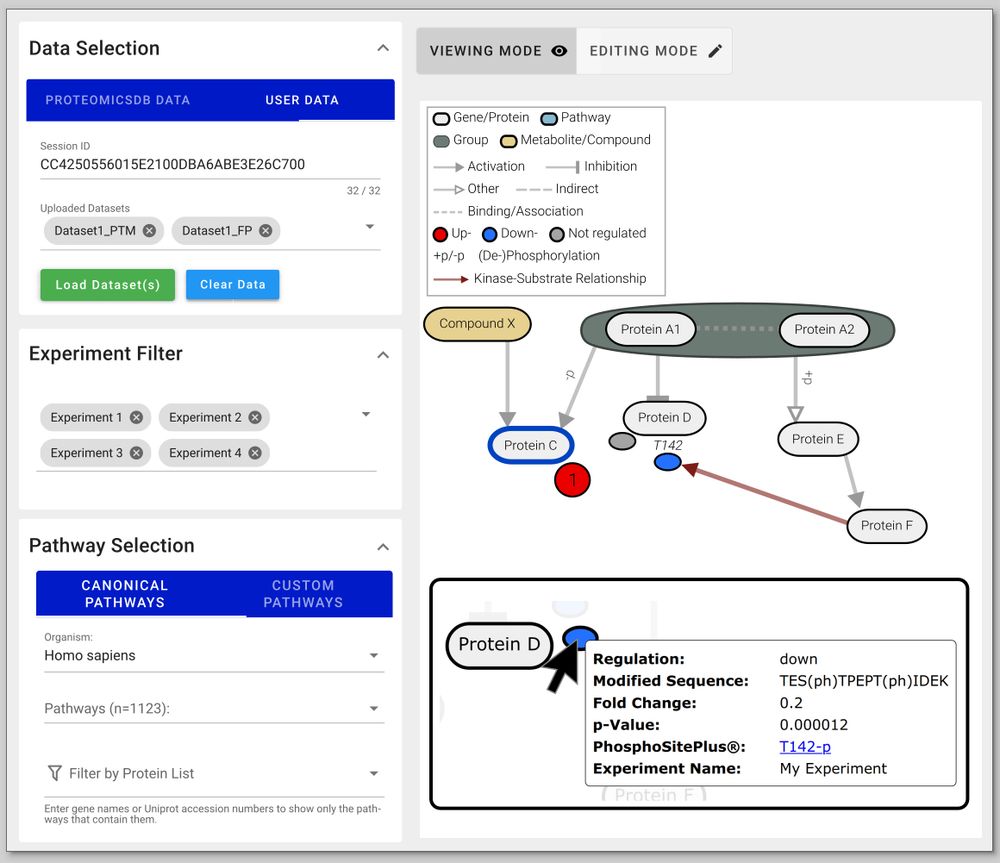
To tackle this, Julian Müller and colleagues have developed PTMNavigator, which is available at ProteomicsDB 💻 www.proteomicsdb.org/analytics/ptmNavigator (2/6)