Kranzusch Lab
@kranzuschlab.bsky.social
Harvard Medical School, Dana Farber Cancer Institute
https://kranzuschlab.med.harvard.edu
https://kranzuschlab.med.harvard.edu
Perfect timing in the field for a beautiful review on NAD+ in bacterial immunity by @hugovaysset.bsky.social and @audeber.bsky.social @cp-molcell.bsky.social
#MicroSky
www.cell.com/molecular-ce...
#MicroSky
www.cell.com/molecular-ce...

October 16, 2025 at 2:58 PM
Perfect timing in the field for a beautiful review on NAD+ in bacterial immunity by @hugovaysset.bsky.social and @audeber.bsky.social @cp-molcell.bsky.social
#MicroSky
www.cell.com/molecular-ce...
#MicroSky
www.cell.com/molecular-ce...
All science arises only because of the shared broader community. I want to especially recognize Rotem Sorek @soreklab.bsky.social Aude Bernheim @audeber.bsky.social our collaborators and the many wonderful labs and scientists in the field bringing together human and bacterial antiviral research!

October 9, 2025 at 4:44 PM
All science arises only because of the shared broader community. I want to especially recognize Rotem Sorek @soreklab.bsky.social Aude Bernheim @audeber.bsky.social our collaborators and the many wonderful labs and scientists in the field bringing together human and bacterial antiviral research!
and amazing people sharing science in our lab. Including members who bravely started exploring bacterial cGAS-like enzymes Aaron @aaronwhiteley.bsky.social Carina @oliveiramann.bsky.social James Eaglesham, Wen Zhou, Ben @benmorehouse.bsky.social Brianna Duncan-Lowey, Apurva Govande, Eric Nieminen

October 9, 2025 at 4:44 PM
and amazing people sharing science in our lab. Including members who bravely started exploring bacterial cGAS-like enzymes Aaron @aaronwhiteley.bsky.social Carina @oliveiramann.bsky.social James Eaglesham, Wen Zhou, Ben @benmorehouse.bsky.social Brianna Duncan-Lowey, Apurva Govande, Eric Nieminen
Thank you to the Blavatnik Family Foundation and The New York Academy of Sciences for honoring our research uniting human innate immunity and bacterial anti-phage defense at @danafarber.bsky.social @harvardmicro.bsky.social @harvard.edu
bit.ly/4pZUDkF
blavatnikawards.org/news/items/t...
bit.ly/4pZUDkF
blavatnikawards.org/news/items/t...

October 9, 2025 at 4:44 PM
Thank you to the Blavatnik Family Foundation and The New York Academy of Sciences for honoring our research uniting human innate immunity and bacterial anti-phage defense at @danafarber.bsky.social @harvardmicro.bsky.social @harvard.edu
bit.ly/4pZUDkF
blavatnikawards.org/news/items/t...
bit.ly/4pZUDkF
blavatnikawards.org/news/items/t...
>18,000 new genomes of giant DNA viruses! An incredible trove of new genes and insights into evolution of host-virus interactions from @fmschu.bsky.social and @jgi.doe.gov
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

September 29, 2025 at 7:10 PM
>18,000 new genomes of giant DNA viruses! An incredible trove of new genes and insights into evolution of host-virus interactions from @fmschu.bsky.social and @jgi.doe.gov
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
NDG is the initial step in our model of a new form of protein conjugation chemistry. Preprints from Doug + Angela Gao and the Bae lab also link NDG to QatABCD defense, highlighting further roles for Q biosynthetic enzymes in immunity
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

September 25, 2025 at 7:16 PM
NDG is the initial step in our model of a new form of protein conjugation chemistry. Preprints from Doug + Angela Gao and the Bae lab also link NDG to QatABCD defense, highlighting further roles for Q biosynthetic enzymes in immunity
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
In phage defense, an N-terminal glycine residue in the substrate protein takes the place of ammonia in canonical Q biosynthetic chemistry to create a protein-nucleobase conjugation modification named N-terminal deazaguanylation or NDG

September 25, 2025 at 7:16 PM
In phage defense, an N-terminal glycine residue in the substrate protein takes the place of ammonia in canonical Q biosynthetic chemistry to create a protein-nucleobase conjugation modification named N-terminal deazaguanylation or NDG
Why defense islands would encode nucleobase biosynthetic machinery was a mystery. We searched in vain for modified nucleotides as signals in CBASS defense… until Doug surprisingly discovered a Q-like base directly conjugated to the N-terminus of a substrate protein!

September 25, 2025 at 7:16 PM
Why defense islands would encode nucleobase biosynthetic machinery was a mystery. We searched in vain for modified nucleotides as signals in CBASS defense… until Doug surprisingly discovered a Q-like base directly conjugated to the N-terminus of a substrate protein!
In addition to ACGU, "Q" is a modified nucleobase required for tRNA maturation in bacteria, plants, and animals. Since 2020, type IV CBASS phage defense islands were known to encode proteins with homology to the enzymes QueC (Q modification) and TGT (Q installation)
www.nature.com/articles/s41...
www.nature.com/articles/s41...

September 25, 2025 at 7:16 PM
In addition to ACGU, "Q" is a modified nucleobase required for tRNA maturation in bacteria, plants, and animals. Since 2020, type IV CBASS phage defense islands were known to encode proteins with homology to the enzymes QueC (Q modification) and TGT (Q installation)
www.nature.com/articles/s41...
www.nature.com/articles/s41...
You’ve heard of ubiquitination, meet deazaguanylation: Doug Wassarman in our lab discovered phage defense pathways have co-opted Q nucleobase biosynthetic enzymes to catalyze a new form of protein conjugation chemistry @science.org
www.science.org/doi/10.1126/...
www.science.org/doi/10.1126/...

September 25, 2025 at 7:16 PM
You’ve heard of ubiquitination, meet deazaguanylation: Doug Wassarman in our lab discovered phage defense pathways have co-opted Q nucleobase biosynthetic enzymes to catalyze a new form of protein conjugation chemistry @science.org
www.science.org/doi/10.1126/...
www.science.org/doi/10.1126/...
Cell death plays important roles in antiviral immunity across all kingdoms of life. Great panel discussion in @natsmb.nature.com from leading experts in the field highlight open mysteries in how and why cell death functions:
www.nature.com/articles/s41...
www.nature.com/articles/s41...

September 18, 2025 at 12:45 PM
Cell death plays important roles in antiviral immunity across all kingdoms of life. Great panel discussion in @natsmb.nature.com from leading experts in the field highlight open mysteries in how and why cell death functions:
www.nature.com/articles/s41...
www.nature.com/articles/s41...
Building on informatics from the labs of Aravind and @teralevin.bsky.social, Yao’s structural analyses enabled her to trace the origins of cGAS-STING and refine a model for evolution of bacterial defense into the immune systems of eukaryotic cells
doi.org/10.1128/jb.0...
doi.org/10.1371/jour...
doi.org/10.1128/jb.0...
doi.org/10.1371/jour...

September 5, 2025 at 10:13 AM
Building on informatics from the labs of Aravind and @teralevin.bsky.social, Yao’s structural analyses enabled her to trace the origins of cGAS-STING and refine a model for evolution of bacterial defense into the immune systems of eukaryotic cells
doi.org/10.1128/jb.0...
doi.org/10.1371/jour...
doi.org/10.1128/jb.0...
doi.org/10.1371/jour...
Yao reasoned single cell eukaryotes may be a model to explain the earliest stages of innate immunity. She discovered in choanoflagellates, the closest living relatives of animals, cGLR (cGAS-like receptor) and STING proteins that are mosaics of features from both bacterial and animal immunity

September 5, 2025 at 10:13 AM
Yao reasoned single cell eukaryotes may be a model to explain the earliest stages of innate immunity. She discovered in choanoflagellates, the closest living relatives of animals, cGLR (cGAS-like receptor) and STING proteins that are mosaics of features from both bacterial and animal immunity
How can we understand the earliest events in evolution of eukaryotic immunity? @yao-li.bsky.social reports incredible molecular fossils of complete bacterial-like operons in eukaryotes that illuminate how animal immunity was first acquired from anti-phage defense
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

September 5, 2025 at 10:13 AM
How can we understand the earliest events in evolution of eukaryotic immunity? @yao-li.bsky.social reports incredible molecular fossils of complete bacterial-like operons in eukaryotes that illuminate how animal immunity was first acquired from anti-phage defense
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Our lab was very happy to help play a part in this story with @reneechang.bsky.social determining new crystal structures that reveal how viral sponge proteins adapt to new host immune signals

August 25, 2025 at 9:06 PM
Our lab was very happy to help play a part in this story with @reneechang.bsky.social determining new crystal structures that reveal how viral sponge proteins adapt to new host immune signals
(2) Equally important, @romihadary.bsky.social and @nitzantal.bsky.social lead creation of an incredible high-throughput assay to rapidly discover the function of new sponge proteins finding as an example the first sponges that block the Pycsar nucleotide immune signals cCMP and cUMP:
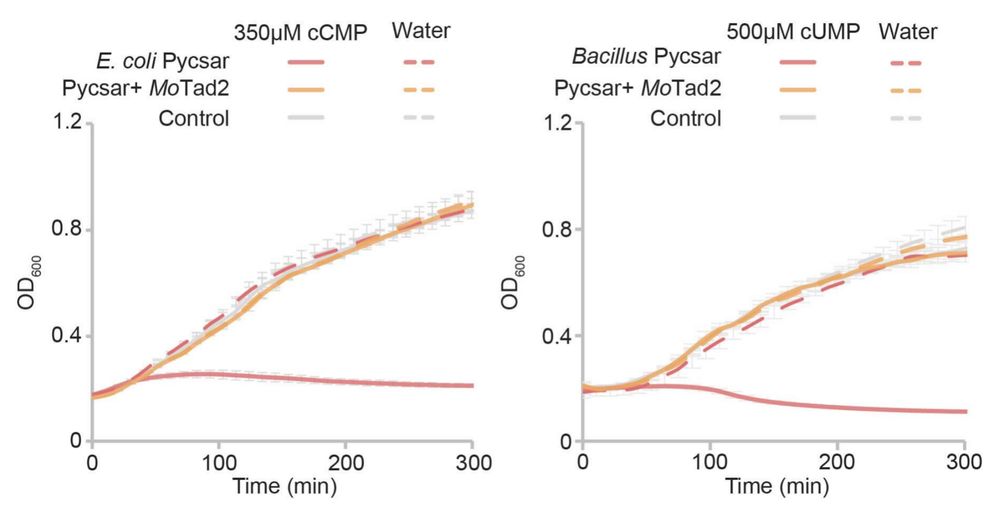
August 25, 2025 at 9:06 PM
(2) Equally important, @romihadary.bsky.social and @nitzantal.bsky.social lead creation of an incredible high-throughput assay to rapidly discover the function of new sponge proteins finding as an example the first sponges that block the Pycsar nucleotide immune signals cCMP and cUMP:
Two major breakthroughs for the nucleotide immune signaling field in pre-print by @romihadary.bsky.social @soreklab.bsky.social www.biorxiv.org/content/10.1...
(1) Beautiful bioinformatic analyses reveal known viral nucleotide sponges are members of enormous protein families ubiquitous in phages
(1) Beautiful bioinformatic analyses reveal known viral nucleotide sponges are members of enormous protein families ubiquitous in phages

August 25, 2025 at 9:06 PM
Two major breakthroughs for the nucleotide immune signaling field in pre-print by @romihadary.bsky.social @soreklab.bsky.social www.biorxiv.org/content/10.1...
(1) Beautiful bioinformatic analyses reveal known viral nucleotide sponges are members of enormous protein families ubiquitous in phages
(1) Beautiful bioinformatic analyses reveal known viral nucleotide sponges are members of enormous protein families ubiquitous in phages
Astounding new structure of a nucleotide immune signal degrading enzyme. The field has found enzymes with this function that look like nucleases, enzymes that look like nucleotide binding proteins, even enzymes that look like proteases... but never a fold like this!
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
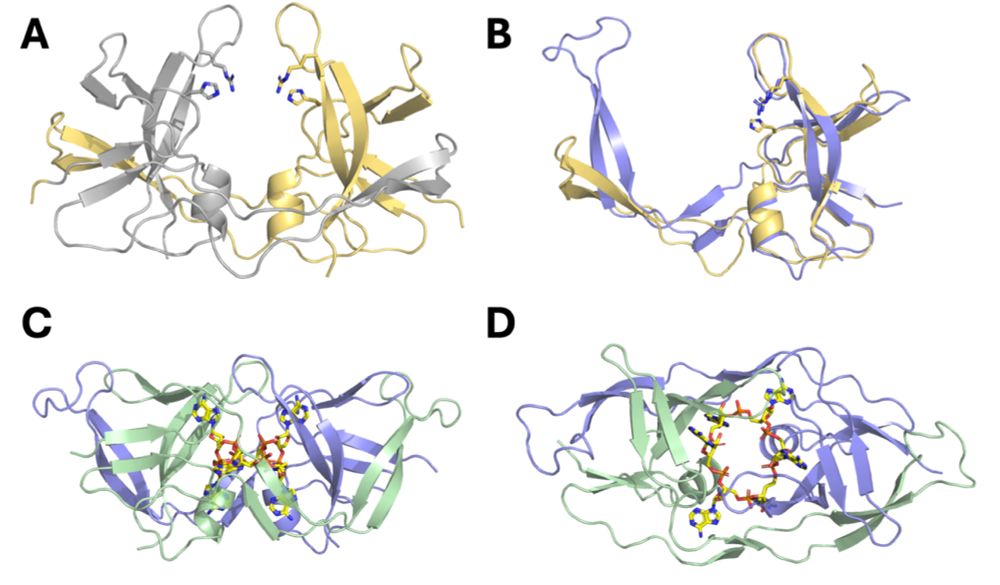
July 18, 2025 at 5:28 PM
Astounding new structure of a nucleotide immune signal degrading enzyme. The field has found enzymes with this function that look like nucleases, enzymes that look like nucleotide binding proteins, even enzymes that look like proteases... but never a fold like this!
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Named after the four-leaf clover architecture of CloA revealed by Sonomi’s cryo-EM structures, Clover senses dTTP to detect viral enzymes that increase nucleotide abundance and is also self-inhibited by the nucleotide signal p3diT that restrains activation to prevent immune-associated toxicity.
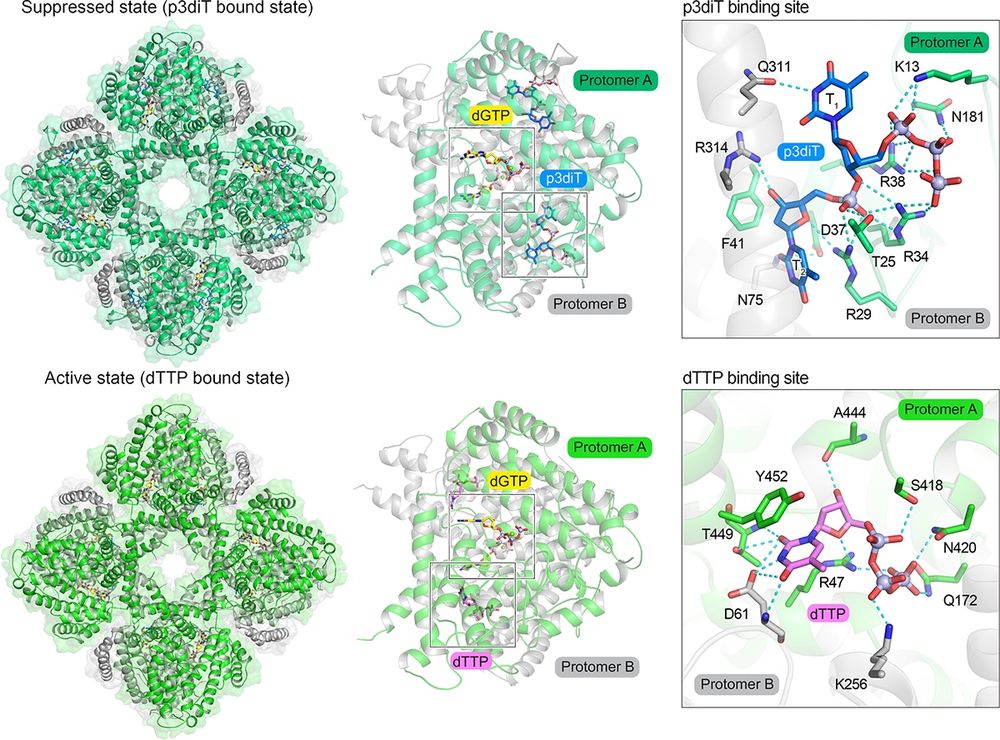
July 17, 2025 at 8:38 PM
Named after the four-leaf clover architecture of CloA revealed by Sonomi’s cryo-EM structures, Clover senses dTTP to detect viral enzymes that increase nucleotide abundance and is also self-inhibited by the nucleotide signal p3diT that restrains activation to prevent immune-associated toxicity.
A new preprint led by Sonomi Yamaguchi in our lab describes a bacterial anti-phage defense system named Clover that uses nucleotide signals to both activate and inhibit host immunity.
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
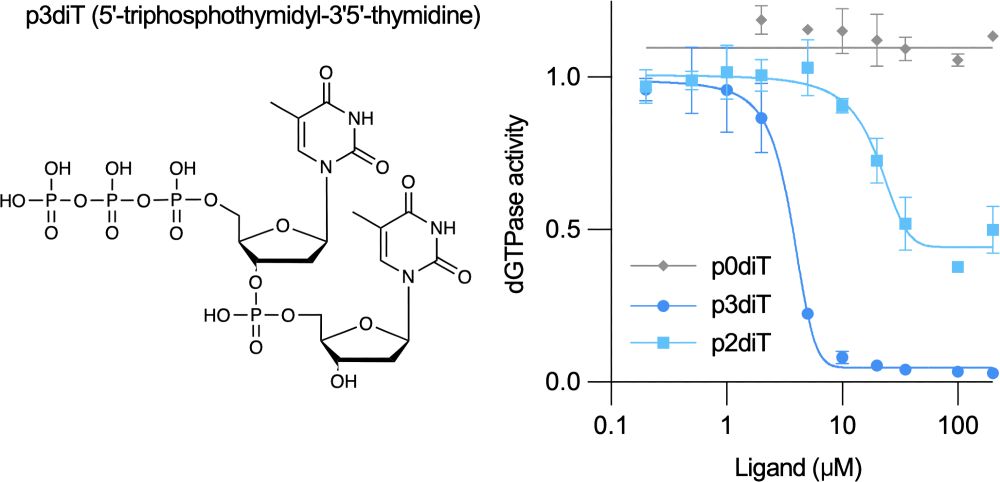
July 17, 2025 at 8:38 PM
A new preprint led by Sonomi Yamaguchi in our lab describes a bacterial anti-phage defense system named Clover that uses nucleotide signals to both activate and inhibit host immunity.
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
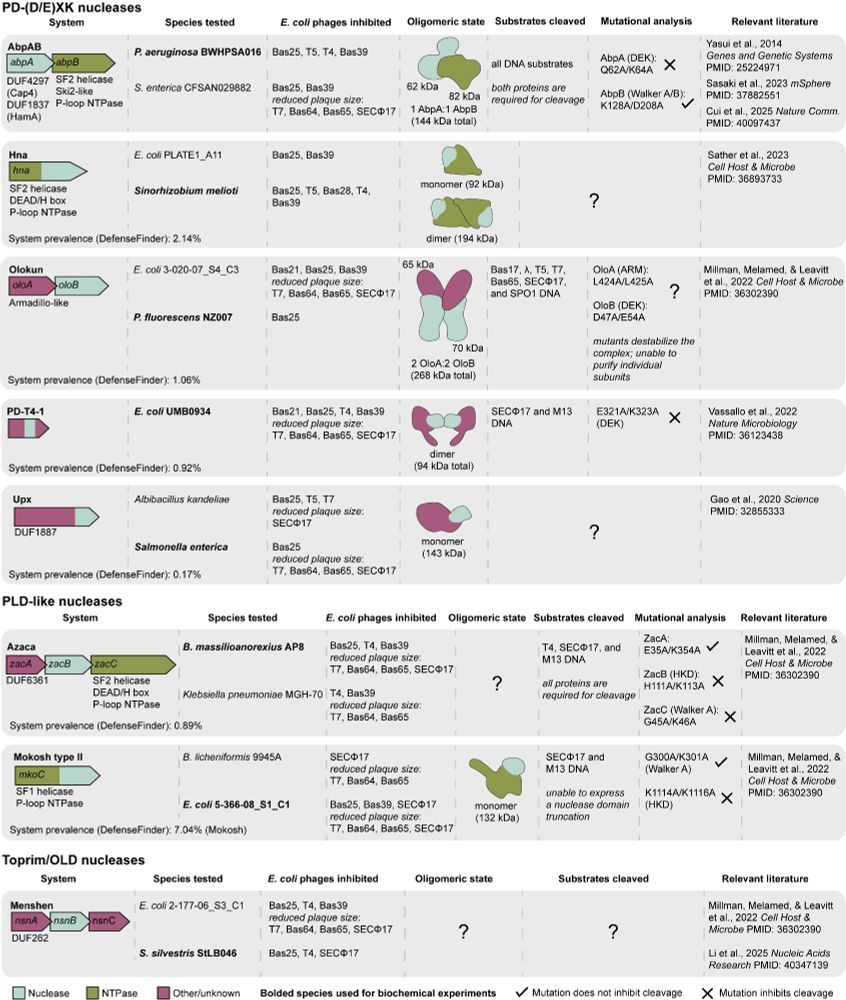
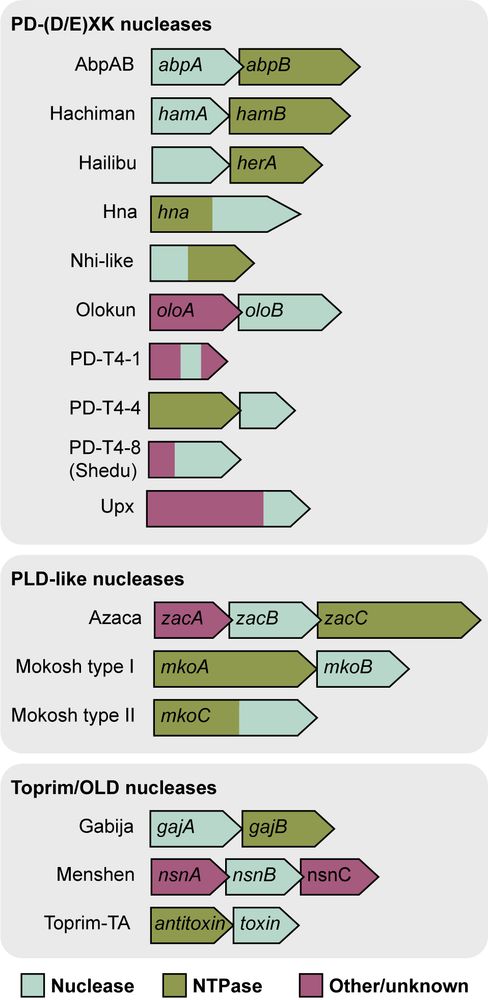
Some nuclease-NTPase systems cleave all DNA targets and others are more specific. Each system requires all complex subunits for target cleavage. Check out Figure 5 for a summary of our data and results in the field that we hope will serve as a guide for others studying these types of systems. (7/7)
July 15, 2025 at 2:25 PM
Some nuclease-NTPase systems cleave all DNA targets and others are more specific. Each system requires all complex subunits for target cleavage. Check out Figure 5 for a summary of our data and results in the field that we hope will serve as a guide for others studying these types of systems. (7/7)
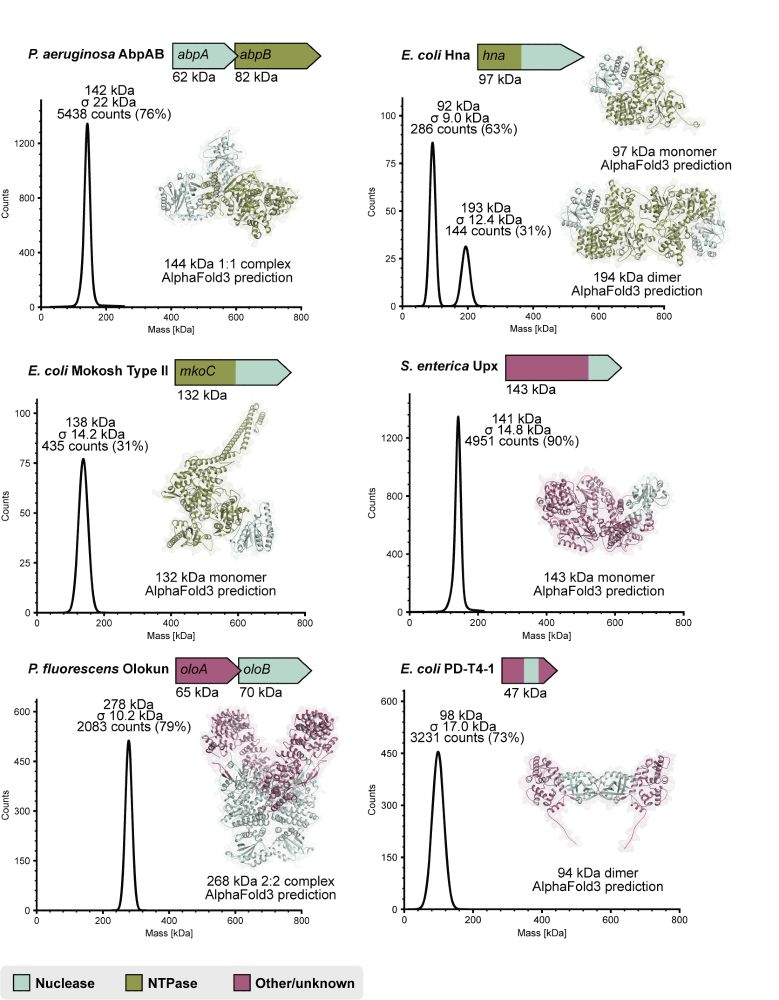
In every system we tested, nuclease and NTPase subunits combine to form large protein complexes. We used mass photometry to create experimentally guided structural models and deep sequencing to define cleavage site specificities. (6/7)
July 15, 2025 at 2:25 PM
In every system we tested, nuclease and NTPase subunits combine to form large protein complexes. We used mass photometry to create experimentally guided structural models and deep sequencing to define cleavage site specificities. (6/7)
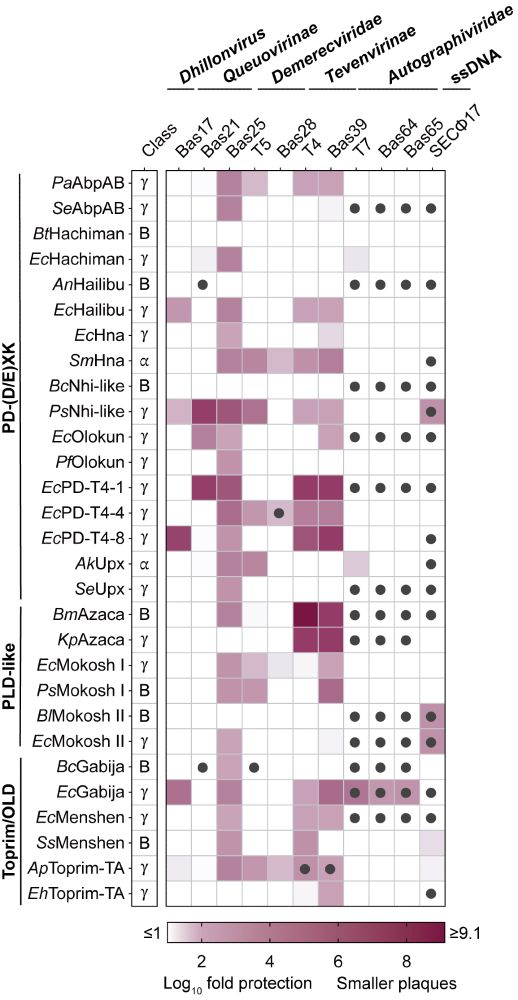
Our large-scale comparative analysis defines shared molecular features of 16 nuclease-NTPase defense systems. Using the @aharms485.bsky.social BASEL phage collection, we show that these systems can exhibit broad anti-phage defense or specific targeting of individual phage families. (5/7)
July 15, 2025 at 2:25 PM
Our large-scale comparative analysis defines shared molecular features of 16 nuclease-NTPase defense systems. Using the @aharms485.bsky.social BASEL phage collection, we show that these systems can exhibit broad anti-phage defense or specific targeting of individual phage families. (5/7)
Nuclease-NTPase systems comprise >10% of recently identified anti-phage defense operons, including AbpAB, Azaca, PD-T4-4, Mokosh, and Menshen. Are other nuclease-NTPase systems similar to Gabija or Hachiman? Can we define the general principles controlling this type of antiviral defense? (4/7)
July 15, 2025 at 2:25 PM
Nuclease-NTPase systems comprise >10% of recently identified anti-phage defense operons, including AbpAB, Azaca, PD-T4-4, Mokosh, and Menshen. Are other nuclease-NTPase systems similar to Gabija or Hachiman? Can we define the general principles controlling this type of antiviral defense? (4/7)
Structures of Gabija from our group and the Zhu, Wang, Fu, and Wei labs explained how nuclease and NTPase subunits assemble into a supramolecular complex to control phage defense, while @doudna-lab.bsky.social structures of Hachiman revealed a 1:1 assembly with distinct DNA cleavage activity. (3/7)
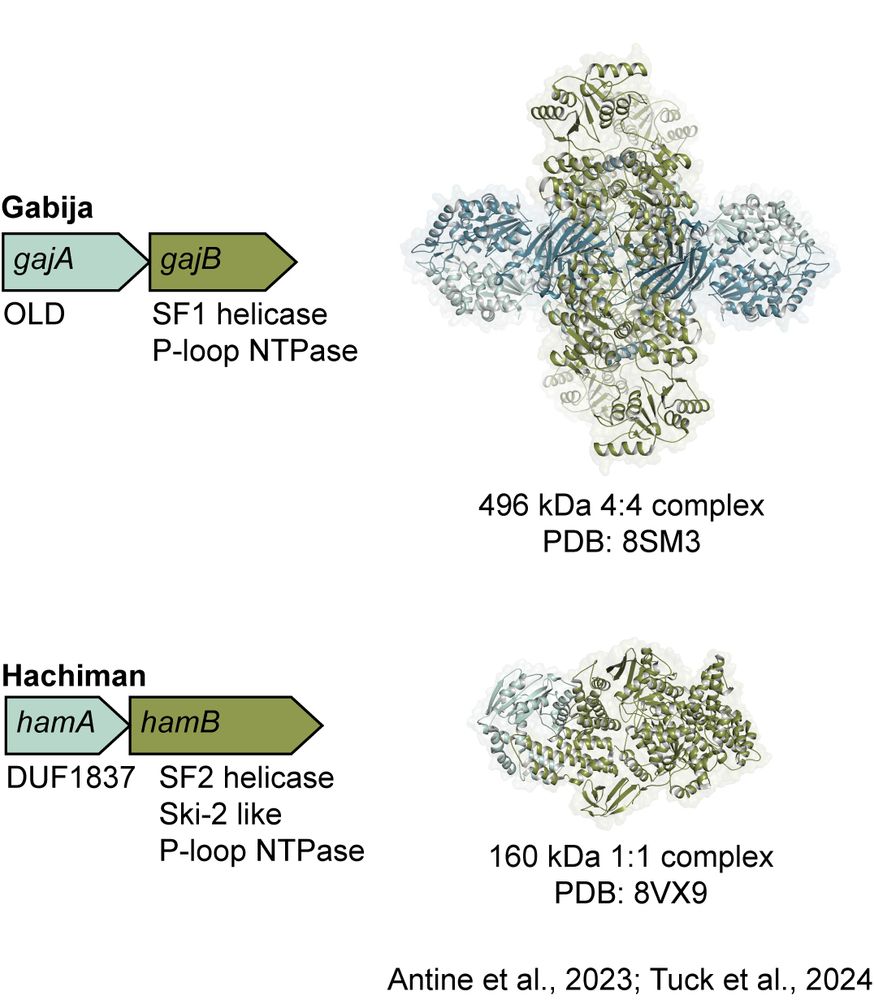
July 15, 2025 at 2:25 PM
Structures of Gabija from our group and the Zhu, Wang, Fu, and Wei labs explained how nuclease and NTPase subunits assemble into a supramolecular complex to control phage defense, while @doudna-lab.bsky.social structures of Hachiman revealed a 1:1 assembly with distinct DNA cleavage activity. (3/7)

