

Neurotransmitters are rude.
Neurotransmitters are rude.
It was a lot of great discussion. But wow, back to back is awful missing one flight, and nearly missing two others by a minute.
It was a lot of great discussion. But wow, back to back is awful missing one flight, and nearly missing two others by a minute.
doi.org/10.1186/s120...

doi.org/10.1186/s120...
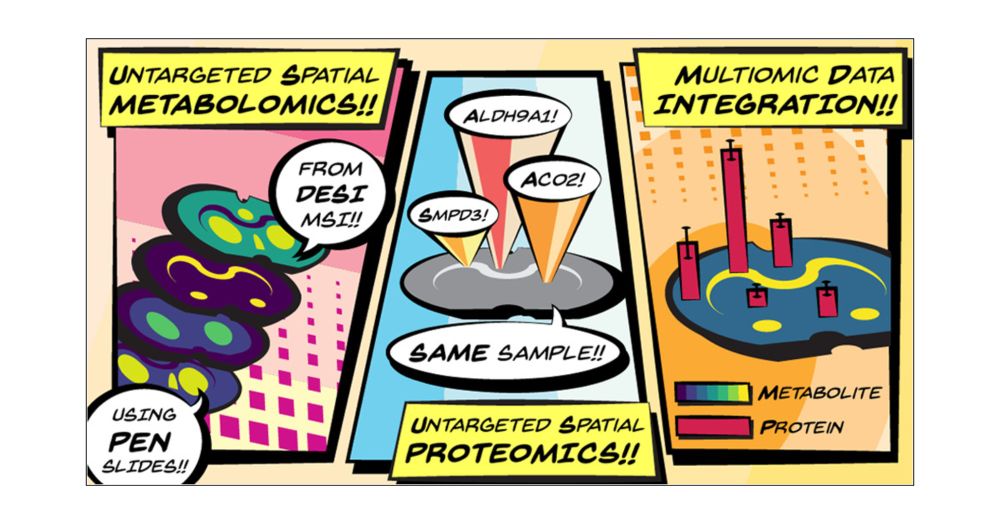
The more advanced the MSI, the more biology unlocked #TeamMassSpec

The more advanced the MSI, the more biology unlocked #TeamMassSpec

While two papers are accepted (not in press), there's a lot of things in review, and much more cool TEAM science is coming. It's been an interesting year, with some shifted priorities.. but here's to a bright (and healthy) 2025!

While two papers are accepted (not in press), there's a lot of things in review, and much more cool TEAM science is coming. It's been an interesting year, with some shifted priorities.. but here's to a bright (and healthy) 2025!

pubs.acs.org/doi/10.1021/...
pubs.acs.org/doi/10.1021/...

Check out this add my colleague Dušan posted for a postdoc here at PNNL. A really amazing opportunity to work with a great team, please share!
@chemjobber.bsky.social #chempostdoc
careers.pnnl.gov/jobs/10140?l...

Check out this add my colleague Dušan posted for a postdoc here at PNNL. A really amazing opportunity to work with a great team, please share!
@chemjobber.bsky.social #chempostdoc
careers.pnnl.gov/jobs/10140?l...

I read should but an 8TB+ SSD for this computer...

I read should but an 8TB+ SSD for this computer...
Maybe will do a little MALDI-7T tomorrow, or maybe more wood.. we shall see..

Maybe will do a little MALDI-7T tomorrow, or maybe more wood.. we shall see..


www.nature.com/articles/s41...




