Counting everything I can, but still appreciating the beauty of qualitative findings.
https://johanneswilbertz.github.io/
Thanks to our collaborators at KSILINK (co-first author Amelie Weiss), LCSB (co-first author Vyron Gorgogietas) and Fraunhofer ITMP.
#Parkinsons #Neurodegeneration #MorphologicalProfiling #iPSC #DrugDiscovery #Mitochondria
Thanks to our collaborators at KSILINK (co-first author Amelie Weiss), LCSB (co-first author Vyron Gorgogietas) and Fraunhofer ITMP.
#Parkinsons #Neurodegeneration #MorphologicalProfiling #iPSC #DrugDiscovery #Mitochondria
This work highlights the power of morphological profiling in human disease-relevant models to uncover multi-phenotype rescue mechanisms.
Data & code: github.com/Ksilink/Notebooks/tree/main/Neuro/PD_MorphProfileScreening

This work highlights the power of morphological profiling in human disease-relevant models to uncover multi-phenotype rescue mechanisms.
Data & code: github.com/Ksilink/Notebooks/tree/main/Neuro/PD_MorphProfileScreening
We propose that mild mitochondrial uncoupling may reduce αSyn burden and oxidative stress, offering a neuroprotective strategy.
Tyrphostin A9 is a tool compound—not a therapeutic—but opens new avenues for drug discovery.
We propose that mild mitochondrial uncoupling may reduce αSyn burden and oxidative stress, offering a neuroprotective strategy.
Tyrphostin A9 is a tool compound—not a therapeutic—but opens new avenues for drug discovery.
Structural analogues (AG879, AG1024) reproduced key effects, suggesting a class effect.
Interestingly, αSyn lowering was not due to proteasomal degradation or transcriptional repression, but likely via autophagy induction.
Structural analogues (AG879, AG1024) reproduced key effects, suggesting a class effect.
Interestingly, αSyn lowering was not due to proteasomal degradation or transcriptional repression, but likely via autophagy induction.
We validated Tyrphostin A9’s effects across multiple assays:
✅ JC-1 dye (Δψm)
✅ ROS & GSH assays
✅ MEA (neuronal firing)
✅ Western blot (αSyn species)
✅ Seahorse (OCR)
✅ Autophagy markers (pAMPK, LC3B, p62)
We validated Tyrphostin A9’s effects across multiple assays:
✅ JC-1 dye (Δψm)
✅ ROS & GSH assays
✅ MEA (neuronal firing)
✅ Western blot (αSyn species)
✅ Seahorse (OCR)
✅ Autophagy markers (pAMPK, LC3B, p62)
Top hits included Tyrphostin A9, a known mitochondrial uncoupler.
It reduced ROS, normalized mitochondrial membrane potential, increased respiration, and lowered αSyn protein levels—without acute toxicity.
Top hits included Tyrphostin A9, a known mitochondrial uncoupler.
It reduced ROS, normalized mitochondrial membrane potential, increased respiration, and lowered αSyn protein levels—without acute toxicity.
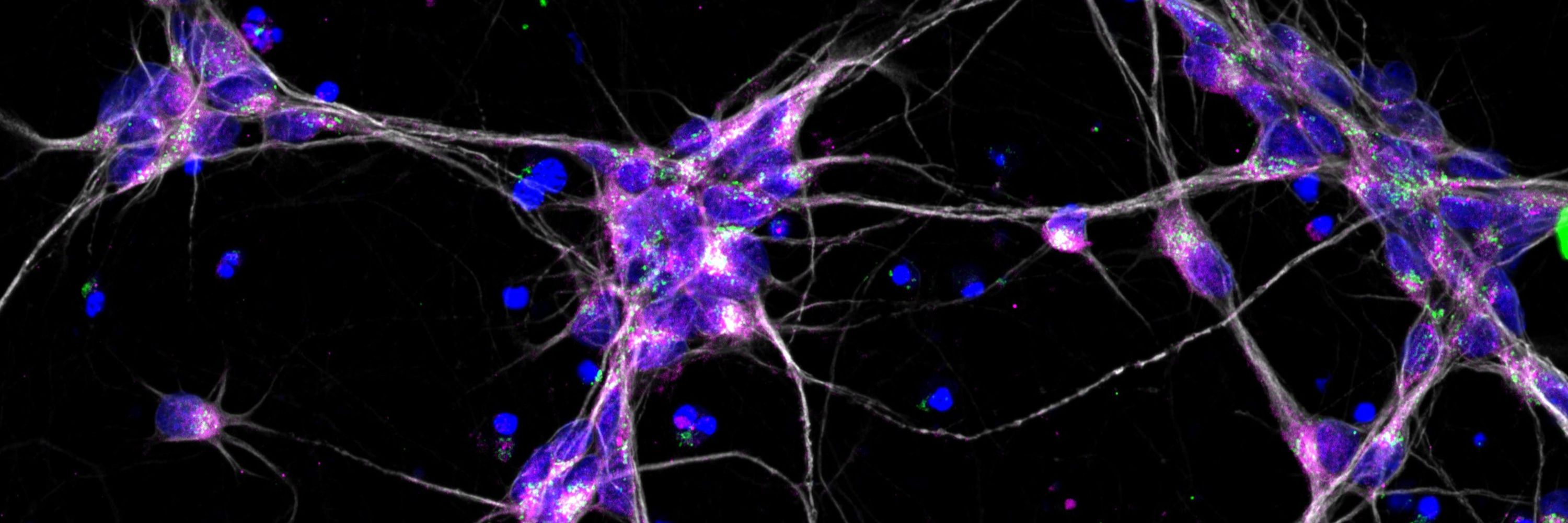
Our approach captured 127 cellular features per well, spanning αSyn distribution, TH expression, mitochondrial health, and more.
Hits were ranked by similarity to isogenic control neurons (SNCA-corr) using Mahalanobis distance and cosine similarity.
Our approach captured 127 cellular features per well, spanning αSyn distribution, TH expression, mitochondrial health, and more.
Hits were ranked by similarity to isogenic control neurons (SNCA-corr) using Mahalanobis distance and cosine similarity.
PD is complex—multiple cellular stressors converge in midbrain dopaminergic neurons.
We modeled PD using patient-derived SNCA triplication neurons (↑αSyn) and screened 1,020 compounds for phenotypic rescue using image-based profiling.
PD is complex—multiple cellular stressors converge in midbrain dopaminergic neurons.
We modeled PD using patient-derived SNCA triplication neurons (↑αSyn) and screened 1,020 compounds for phenotypic rescue using image-based profiling.
Gift link for #TCTeAC:
rdcu.be/eqPpi
www.nature.com/articles/s41...
Gift link for #TCTeAC:
rdcu.be/eqPpi
www.nature.com/articles/s41...

For a general term like "in this study", the results for 2023-2024 were exactly on the trendline based on the 2014-2022 data. So probably, there is no reason to believe that publication numbers rose in general after 2022, but only certain word frequencies changed.

For a general term like "in this study", the results for 2023-2024 were exactly on the trendline based on the 2014-2022 data. So probably, there is no reason to believe that publication numbers rose in general after 2022, but only certain word frequencies changed.
Unexpectedly, there was a drop in abstracts using widespread words like "our results" or "we show" after 2022. This might indicate the use of LLMs by researchers, as LLM-generated text often uses passive rather than active language ("this shows" versus "we show").


Unexpectedly, there was a drop in abstracts using widespread words like "our results" or "we show" after 2022. This might indicate the use of LLMs by researchers, as LLM-generated text often uses passive rather than active language ("this shows" versus "we show").
Some words are clearly more frequently found in biomedical literature after 2022, such as "pertinent", "treasure trove" or "shedding light". This could be due to overall publication numbers increasing for LLM-unrelated reasons.



Some words are clearly more frequently found in biomedical literature after 2022, such as "pertinent", "treasure trove" or "shedding light". This could be due to overall publication numbers increasing for LLM-unrelated reasons.

