
💻🧬 Using data science to analyze big bio-data.
🇪🇺 Part of the @embltrec.bsky.social expedition and interested in 🦠 (metaG)

In this study, we highlight that LDGs are not universal in marine microbiomes but reflect lineage-specific ecological strategies and environmental responses.
doi.org/10.1101/2025...
Metalog: curated and harmonised contextual data for global metagenomics samples
now out in @narjournal.bsky.social
academic.oup.com/nar/advance-...

Metalog: curated and harmonised contextual data for global metagenomics samples
now out in @narjournal.bsky.social
academic.oup.com/nar/advance-...
In this study, we highlight that LDGs are not universal in marine microbiomes but reflect lineage-specific ecological strategies and environmental responses.
doi.org/10.1101/2025...

In this study, we highlight that LDGs are not universal in marine microbiomes but reflect lineage-specific ecological strategies and environmental responses.
doi.org/10.1101/2025...

In this study, we highlight that LDGs are not universal in marine microbiomes but reflect lineage-specific ecological strategies and environmental responses.
doi.org/10.1101/2025...
Check out our newest preprint!
We show that latitudinal diversity gradients are taxon-specific, reflecting differing ecological strategies and their responses to environmental gradients
www.biorxiv.org/content/10.1...

Check out our newest preprint!
We show that latitudinal diversity gradients are taxon-specific, reflecting differing ecological strategies and their responses to environmental gradients
www.biorxiv.org/content/10.1...
At EMBL, we train young scientists to become skilled and creative future leaders in academia and other sectors. Join us and get a head start on your career in life sciences!
bit.ly/47mmiFF

At EMBL, we train young scientists to become skilled and creative future leaders in academia and other sectors. Join us and get a head start on your career in life sciences!
bit.ly/47mmiFF

Proud to have contributed to Metalog, the latest @borklab.bsky.social resource:
www.biorxiv.org/content/10.1...
#microsky #microbiome
Proud to have contributed to Metalog, the latest @borklab.bsky.social resource:
www.biorxiv.org/content/10.1...
#microsky #microbiome

#microsky
#microsky

#MicroSky 🖥️🧬🦠
www.biorxiv.org/content/10.1...
In this study, we present the largest systematic analysis of microbiome structure and function, integrating 85K uniformly processed metagenomes from diverse habitats worldwide.
@podlesny.bsky.social @jonas-bio.bsky.social @borklab.bsky.social

#MicroSky 🖥️🧬🦠
#Microbiology #Metagenomics #HGT #ARG
www.biorxiv.org/content/10.1...
In this study, we present the largest systematic analysis of microbiome structure and function, integrating 85K uniformly processed metagenomes from diverse habitats worldwide.
@podlesny.bsky.social @jonas-bio.bsky.social @borklab.bsky.social

#Microbiology #Metagenomics #HGT #ARG
We explored the diversity, temporal dynamics and host-associations of Arctic #viruses over a four year time-series using long-read #metagenomics (and their global prevalence and distribution).
www.nature.com/articles/s41...
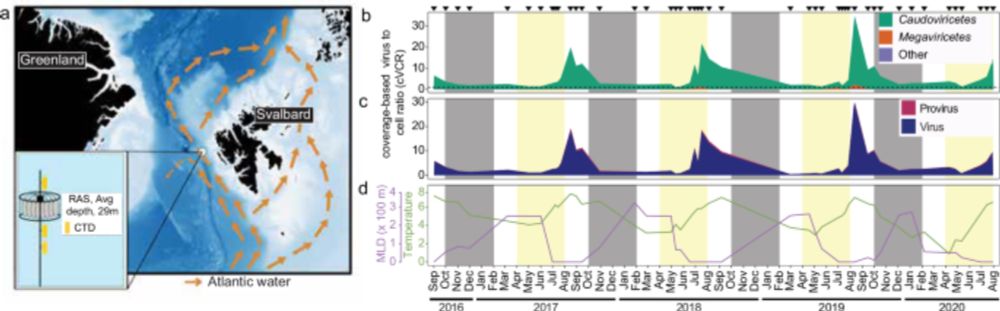
We explored the diversity, temporal dynamics and host-associations of Arctic #viruses over a four year time-series using long-read #metagenomics (and their global prevalence and distribution).
www.nature.com/articles/s41...
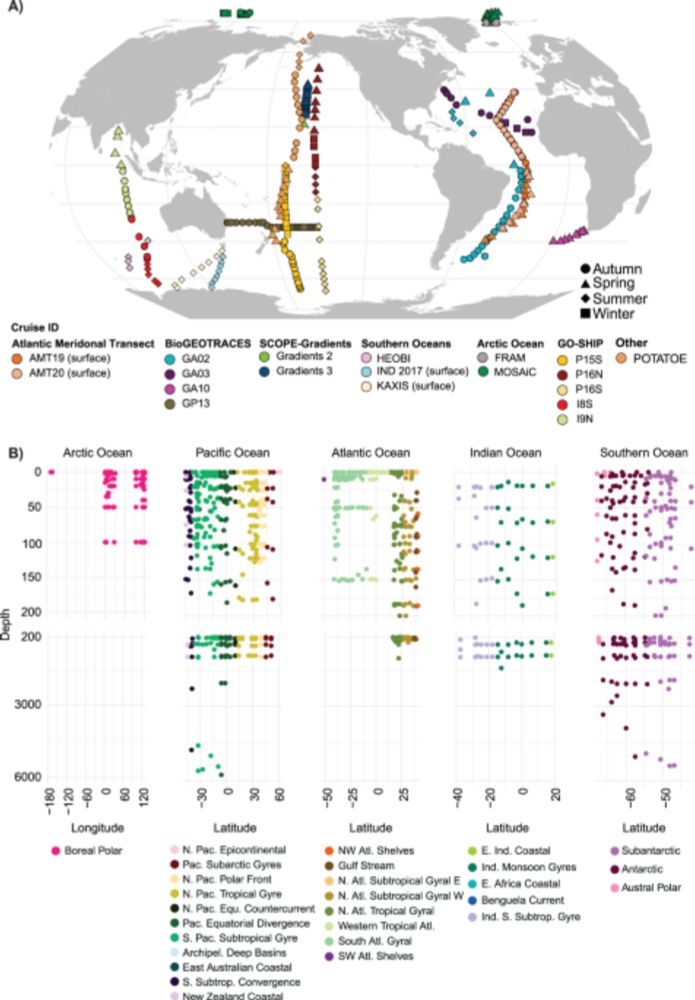
Led by
@vishnuprasoodanan.bsky.social & @omaistrenko.bsky.social , we asked a question (almost) as old as microbiology: how many prokaryotic species exist on Earth? More specifically, how much diversity is "hiding" in existing metagenomic data?
A 🧵.
Led by
@vishnuprasoodanan.bsky.social & @omaistrenko.bsky.social , we asked a question (almost) as old as microbiology: how many prokaryotic species exist on Earth? More specifically, how much diversity is "hiding" in existing metagenomic data?
A 🧵.
Ever wondered how to best quantify cell-cell neighbor preferences in tissues?
We compared 9+ neighbor preference (NEP) methods for analysing spatial omics data and propose a novel approach that combines the most relevant analysis features which we call COZI 🔬✨
Read more: doi.org/10.1101/2025...
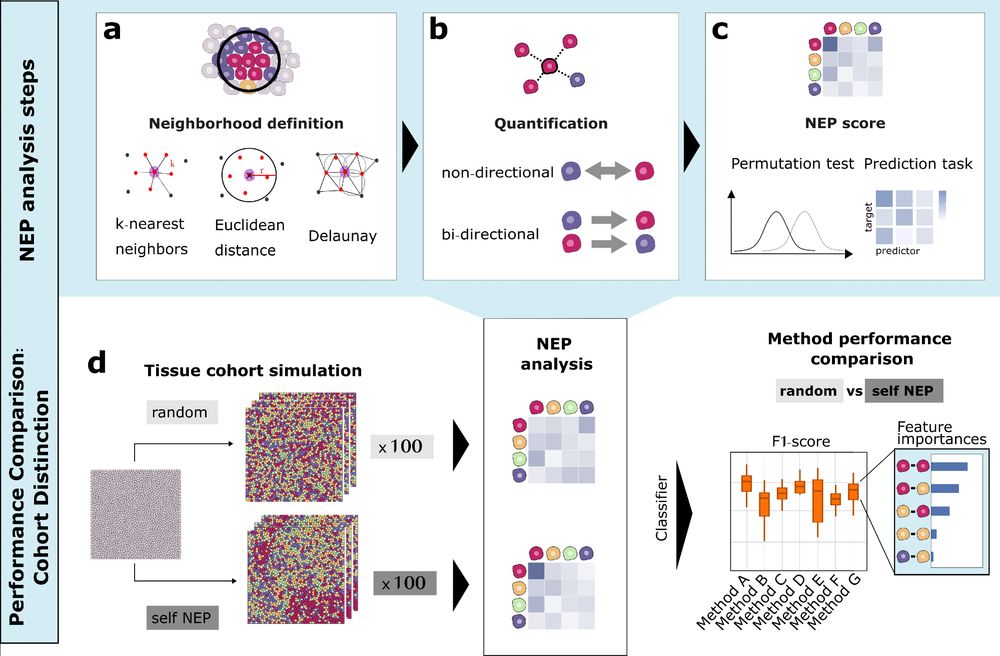
Ever wondered how to best quantify cell-cell neighbor preferences in tissues?
We compared 9+ neighbor preference (NEP) methods for analysing spatial omics data and propose a novel approach that combines the most relevant analysis features which we call COZI 🔬✨
Read more: doi.org/10.1101/2025...
Fantastic teamwork by #AWI @mpimarinemicrobio.bsky.social @hhu.bsky.social & many others 🙂

Fantastic teamwork by #AWI @mpimarinemicrobio.bsky.social @hhu.bsky.social & many others 🙂
So happy that this study is out. Big thanks to co-first author @bellahda.bsky.social and all the collaborators!
www.science.org/doi/10.1126/...
@science.org @mblscience.bsky.social @simonsfoundation.org #deeplife #microbiome
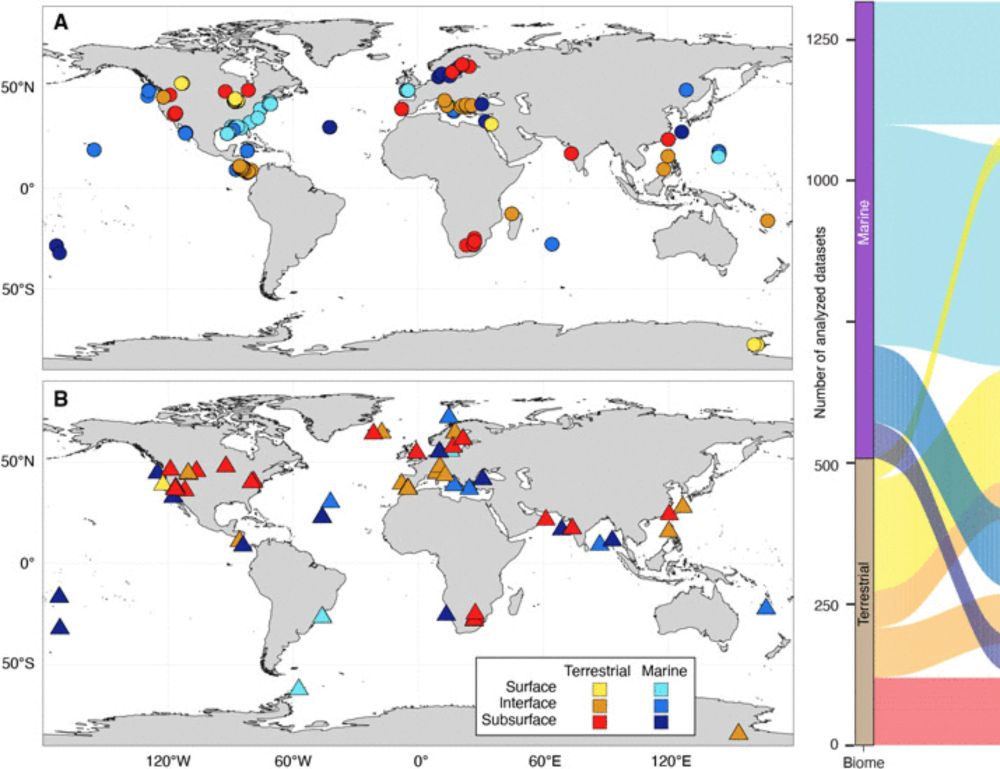
So happy that this study is out. Big thanks to co-first author @bellahda.bsky.social and all the collaborators!
www.science.org/doi/10.1126/...
@science.org @mblscience.bsky.social @simonsfoundation.org #deeplife #microbiome

