

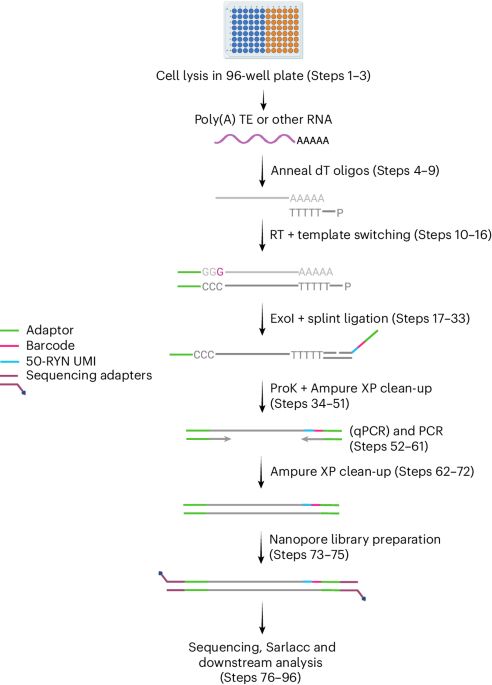
www.biorxiv.org/content/10.1...
Read below for a few highlights...
www.biorxiv.org/content/10.1...
Read below for a few highlights...
She reveals how SMCHD1 dynamically engages chromatin, including the inactive X chromosome, to maintain gene silencing.
www.biorxiv.org/content/10.1...

She reveals how SMCHD1 dynamically engages chromatin, including the inactive X chromosome, to maintain gene silencing.
www.biorxiv.org/content/10.1...

🧬 Single cell methylome encodes cell state & clonal identity
🔨 EPI-Clone reads out both (+mutations, +RNA) at scale
🩸 Clonal expansions of HSCs are universal from age 50, not driven by CH mutations
doi.org/10.1038/s415...
🧵
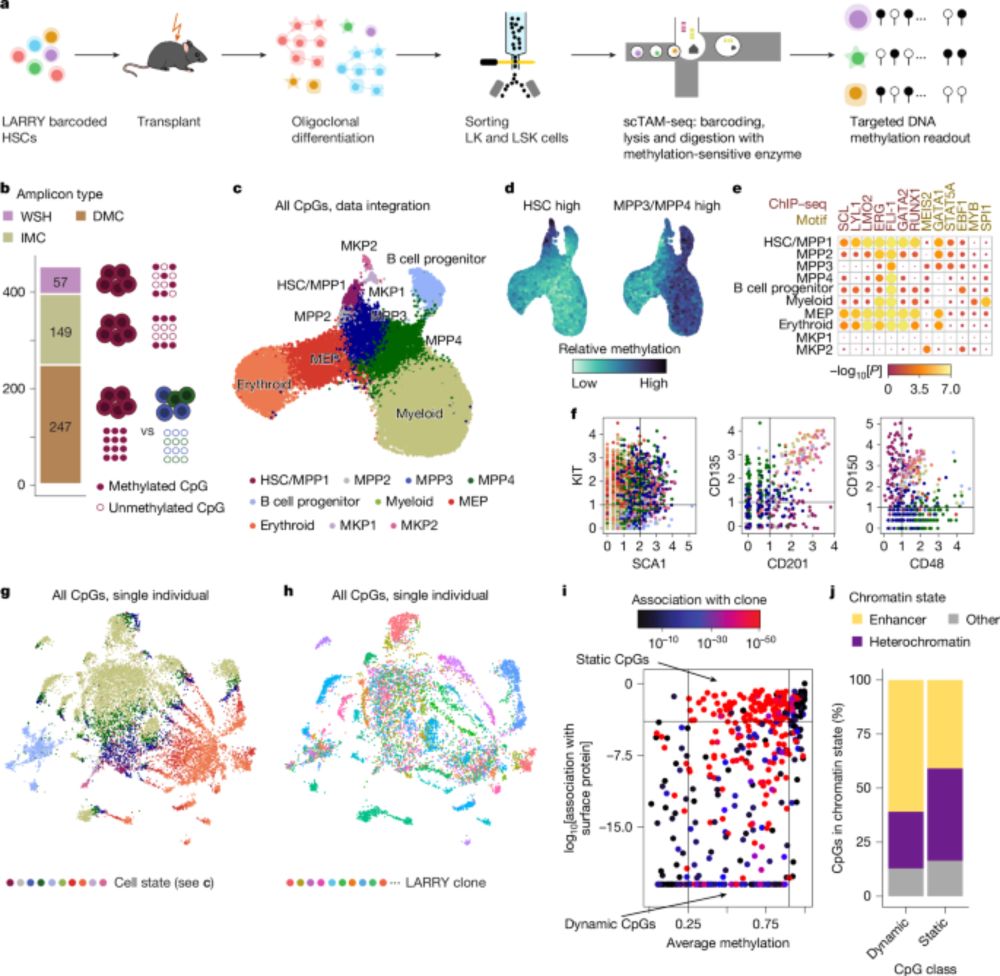


Read all about it this really cool work below ⬇️
🧬🩸 screen of fully synthetic enhancers in blood progenitors
🤖 AI that creates new cell state specific enhancers
🔍 negative synergies between TFs lead to specificity!
www.cell.com/cell/fulltex...
🧵

Read all about it this really cool work below ⬇️

A 🧵 below 👇🏼




