Joe Marsh
@jmarshlab.bsky.social
Reposted by Joe Marsh
Happy to share that 𝚊𝚌𝚖𝚐𝚜𝚌𝚊𝚕𝚎𝚛 is now on CRAN! 🎉
This means long-term stability and easy installation with:
𝚒𝚗𝚜𝚝𝚊𝚕𝚕.𝚙𝚊𝚌𝚔𝚊𝚐𝚎𝚜('𝚊𝚌𝚖𝚐𝚜𝚌𝚊𝚕𝚎𝚛')
🗞️ doi.org/10.1093/bioi...
#rstats #acmg #varianteffect #MAVEs #VEPs #genomics
This means long-term stability and easy installation with:
𝚒𝚗𝚜𝚝𝚊𝚕𝚕.𝚙𝚊𝚌𝚔𝚊𝚐𝚎𝚜('𝚊𝚌𝚖𝚐𝚜𝚌𝚊𝚕𝚎𝚛')
🗞️ doi.org/10.1093/bioi...
#rstats #acmg #varianteffect #MAVEs #VEPs #genomics
New on CRAN: acmgscaler (1.0.0). View at https://CRAN.R-project.org/package=acmgscaler

acmgscaler: Variant Effect Calibration to ACMG/AMP Evidence Strength
Provides a function to calibrate variant effect scores against evidence strength categories defined by the American College of Medical Genetics and Genomics (ACMG) and the Association for Molecular Pathology (AMP) guidelines. The method computes likelihood ratios of pathogenicity via kernel density estimation of pathogenic and benign score distributions, and derives score intervals corresponding to ACMG/AMP evidence levels. This enables researchers and clinical geneticists to interpret functional and computational variant scores in a reproducible and standardised manner. For details, see Badonyi and Marsh (2025) <<a href="https://doi.org/10.1093%2Fbioinformatics%2Fbtaf503" target="_top">doi:10.1093/bioinformatics/btaf503</a>>.
CRAN.R-project.org
September 24, 2025 at 6:31 AM
Happy to share that 𝚊𝚌𝚖𝚐𝚜𝚌𝚊𝚕𝚎𝚛 is now on CRAN! 🎉
This means long-term stability and easy installation with:
𝚒𝚗𝚜𝚝𝚊𝚕𝚕.𝚙𝚊𝚌𝚔𝚊𝚐𝚎𝚜('𝚊𝚌𝚖𝚐𝚜𝚌𝚊𝚕𝚎𝚛')
🗞️ doi.org/10.1093/bioi...
#rstats #acmg #varianteffect #MAVEs #VEPs #genomics
This means long-term stability and easy installation with:
𝚒𝚗𝚜𝚝𝚊𝚕𝚕.𝚙𝚊𝚌𝚔𝚊𝚐𝚎𝚜('𝚊𝚌𝚖𝚐𝚜𝚌𝚊𝚕𝚎𝚛')
🗞️ doi.org/10.1093/bioi...
#rstats #acmg #varianteffect #MAVEs #VEPs #genomics
Reposted by Joe Marsh
1/8 Our new paper in Nature Communications explores how often pathogenic missense variants cause disease through loss-of-function (LOF), gain-of-function (GOF), or dominant-negative (DN) effects.
📄 nature.com/articles/s41...
📄 nature.com/articles/s41...
Find out more about a new online tool which allows researchers and clinicians to estimate likely disease mechanisms for sets of mutations, supporting better diagnosis and personalised medicine 👉 edin.ac/3W5ykw3
@mbadonyi.bsky.social @jmarshlab.bsky.social
@mbadonyi.bsky.social @jmarshlab.bsky.social

New research classifies mutant proteins behind genetic diseases | Institute of Genetics and Cancer | Institute of Genetics and Cancer
A new score classifying dominant disease-causing mutations has found that just over half act via loss of function, with gain-of-function and dominant-negative mechanisms making up the rest.
edin.ac
September 25, 2025 at 2:14 PM
1/8 Our new paper in Nature Communications explores how often pathogenic missense variants cause disease through loss-of-function (LOF), gain-of-function (GOF), or dominant-negative (DN) effects.
📄 nature.com/articles/s41...
📄 nature.com/articles/s41...
Happy to see this out, check out our paper here: www.nature.com/articles/s41...
September 25, 2025 at 2:01 PM
Happy to see this out, check out our paper here: www.nature.com/articles/s41...
New paper out today in PLOS Comp Biol:
journals.plos.org/ploscompbiol...
Intrinsically disordered regions make variant prediction deceptively easy for benign changes but very hard for pathogenic ones. Our work shows why current tools struggle here, and why disorder-aware approaches are needed.
journals.plos.org/ploscompbiol...
Intrinsically disordered regions make variant prediction deceptively easy for benign changes but very hard for pathogenic ones. Our work shows why current tools struggle here, and why disorder-aware approaches are needed.

Assessing variant effect predictors and disease mechanisms in intrinsically disordered proteins
Author summary Some parts of proteins, known as intrinsically disordered regions, do not fold into fixed shapes. Instead, they stay flexible and play key roles in controlling how cells work, often by ...
journals.plos.org
August 19, 2025 at 9:33 PM
New paper out today in PLOS Comp Biol:
journals.plos.org/ploscompbiol...
Intrinsically disordered regions make variant prediction deceptively easy for benign changes but very hard for pathogenic ones. Our work shows why current tools struggle here, and why disorder-aware approaches are needed.
journals.plos.org/ploscompbiol...
Intrinsically disordered regions make variant prediction deceptively easy for benign changes but very hard for pathogenic ones. Our work shows why current tools struggle here, and why disorder-aware approaches are needed.
Reposted by Joe Marsh
We’ve updated the acmgscaler manuscript following reviewer and community feedback.
The R package now has a single calibrate() function, and the Colab interface is easier to use.
📄 Manuscript: www.biorxiv.org/content/10.1...
🧪 Colab: edin.ac/4mjzijp
#rstats @theacmg.bsky.social
The R package now has a single calibrate() function, and the Colab interface is easier to use.
📄 Manuscript: www.biorxiv.org/content/10.1...
🧪 Colab: edin.ac/4mjzijp
#rstats @theacmg.bsky.social

acmgscaler: An R package and Colab for standardised gene-level variant effect score calibration within the ACMG/AMP framework
A genome-wide variant effect calibration method was recently developed under the guidelines of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology (ACMG/A...
www.biorxiv.org
August 15, 2025 at 8:35 AM
We’ve updated the acmgscaler manuscript following reviewer and community feedback.
The R package now has a single calibrate() function, and the Colab interface is easier to use.
📄 Manuscript: www.biorxiv.org/content/10.1...
🧪 Colab: edin.ac/4mjzijp
#rstats @theacmg.bsky.social
The R package now has a single calibrate() function, and the Colab interface is easier to use.
📄 Manuscript: www.biorxiv.org/content/10.1...
🧪 Colab: edin.ac/4mjzijp
#rstats @theacmg.bsky.social
New preprint from our group - Ben has done some great work trying to understand why computational predictors and MAVEs agree or disagree when scoring the impacts of single amino acid substitutions
Check out our latest preprint: "Why variant effect predictors and multiplexed assays agree and disagree". Now available on BioRxiv www.biorxiv.org/content/10.1... @marshlab.bsky.social @uoe-igc.bsky.social

Why variant effect predictors and multiplexed assays agree and disagree
Computational variant effect predictors (VEPs) and multiplexed assays of variant effect (MAVEs) are two key tools for assessing the functional consequences of genetic variants. While their outputs are...
www.biorxiv.org
August 4, 2025 at 8:38 PM
New preprint from our group - Ben has done some great work trying to understand why computational predictors and MAVEs agree or disagree when scoring the impacts of single amino acid substitutions
Reposted by Joe Marsh
GWAS to mechanism: when non-coding is coding. Beautiful insightful science from @gweykopf.bsky.social @simonbiddie.bsky.social Joe Marsh and many colleagues. @uoe-igc.bsky.social @cmvm-edinburghuni.bsky.social www.biorxiv.org/content/10.1...
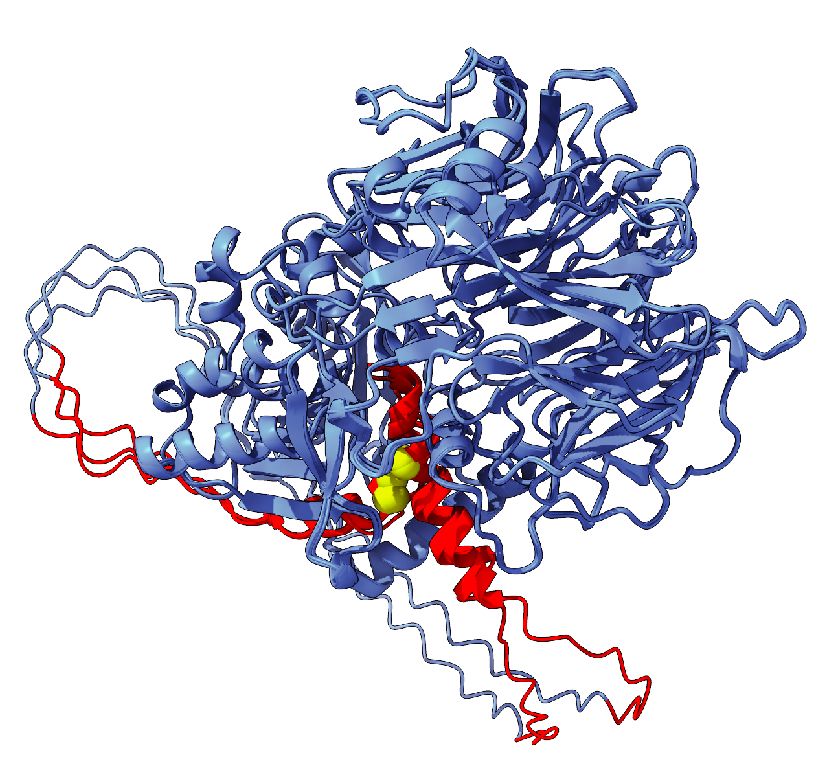
July 28, 2025 at 9:50 AM
GWAS to mechanism: when non-coding is coding. Beautiful insightful science from @gweykopf.bsky.social @simonbiddie.bsky.social Joe Marsh and many colleagues. @uoe-igc.bsky.social @cmvm-edinburghuni.bsky.social www.biorxiv.org/content/10.1...
Reposted by Joe Marsh
Pleased to share our latest work and the first manuscript from the Degron Tagging Cluster in the MRC National Mouse Genetics Network. If you work with protein tags, particularly in tissue biology models, this should be of interest:
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
www.biorxiv.org
June 19, 2025 at 8:56 PM
Pleased to share our latest work and the first manuscript from the Degron Tagging Cluster in the MRC National Mouse Genetics Network. If you work with protein tags, particularly in tissue biology models, this should be of interest:
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Excited to share this new method for gene-level calibration of MAVE and VEP scores that Mihaly has been working so hard on!
We've developed a method to align genetic variant effect scores with ACMG/AMP classification criteria. It has two key advantages: (1) no assumptions about score distributions, and (2) consistent outputs without user tuning.
acmgscaler: An R package and Colab for standardised gene-level variant effect score calibration within the ACMG/AMP framework https://www.biorxiv.org/content/10.1101/2025.05.16.654507v1
May 22, 2025 at 9:16 AM
Excited to share this new method for gene-level calibration of MAVE and VEP scores that Mihaly has been working so hard on!
Reposted by Joe Marsh
acmgscaler: An R package and Colab for standardised gene-level variant effect score calibration within the ACMG/AMP framework https://www.biorxiv.org/content/10.1101/2025.05.16.654507v1
May 21, 2025 at 6:48 PM
acmgscaler: An R package and Colab for standardised gene-level variant effect score calibration within the ACMG/AMP framework https://www.biorxiv.org/content/10.1101/2025.05.16.654507v1
Reposted by Joe Marsh
We are hiring!
Want to join my new group at the amazing @uoe-igc.bsky.social and perform ground-breaking studies in reproductive genomics and genomic medicine as a computational genomicist?
Please DM me to discuss this, I will be attending #ESHG2025
elxw.fa.em3.oraclecloud.com/hcmUI/Candid...
Want to join my new group at the amazing @uoe-igc.bsky.social and perform ground-breaking studies in reproductive genomics and genomic medicine as a computational genomicist?
Please DM me to discuss this, I will be attending #ESHG2025
elxw.fa.em3.oraclecloud.com/hcmUI/Candid...
Lecturer in Computational Genomics
Establishing an innovative research line as Lecturer in Computational Genomics related to reproductive disorders and genomic medicine. This Lecturer post is full-time (35 hours per week); however, we ...
elxw.fa.em3.oraclecloud.com
May 16, 2025 at 12:45 PM
We are hiring!
Want to join my new group at the amazing @uoe-igc.bsky.social and perform ground-breaking studies in reproductive genomics and genomic medicine as a computational genomicist?
Please DM me to discuss this, I will be attending #ESHG2025
elxw.fa.em3.oraclecloud.com/hcmUI/Candid...
Want to join my new group at the amazing @uoe-igc.bsky.social and perform ground-breaking studies in reproductive genomics and genomic medicine as a computational genomicist?
Please DM me to discuss this, I will be attending #ESHG2025
elxw.fa.em3.oraclecloud.com/hcmUI/Candid...
Reposted by Joe Marsh
Very excited to see our recent preprint covered here! @mbadonyi.bsky.social
So, how many genetic diseases come down to good ol’ loss-of-function in the targeted protein?
Your estimate is probably too high:
Your estimate is probably too high:

Mutant Proteins Classified
www.science.org
May 8, 2025 at 10:04 AM
Very excited to see our recent preprint covered here! @mbadonyi.bsky.social
Reposted by Joe Marsh
Read more about this study by @jmarshlab.bsky.social 👇
April 30, 2025 at 2:40 PM
Read more about this study by @jmarshlab.bsky.social 👇
Reposted by Joe Marsh
Mutational Scanning helps guide precision medicine! But how does it work? 🤔 Check out this Introduction to Deep Mutational Scanning (Animation) @uwgenome.bsky.social www.youtube.com/watch?v=NRKj...

Introduction to Deep Mutational Scanning (Animation)
YouTube video by Variant Effects
www.youtube.com
April 24, 2025 at 5:07 PM
Mutational Scanning helps guide precision medicine! But how does it work? 🤔 Check out this Introduction to Deep Mutational Scanning (Animation) @uwgenome.bsky.social www.youtube.com/watch?v=NRKj...
Reposted by Joe Marsh
The guidelines "aim to streamline VEP development, sharing, and evaluation by tackling data availability, interpretability, transparency, and circularity." Benjamin J. Livesey, @jmarshlab.bsky.social et al
genomebiology.biomedcentral.com/articles/10....
genomebiology.biomedcentral.com/articles/10....

Guidelines for releasing a variant effect predictor - Genome Biology
Computational methods for assessing the likely impacts of mutations, known as variant effect predictors (VEPs), are widely used in the assessment and interpretation of human genetic variation, as well...
genomebiology.biomedcentral.com
April 24, 2025 at 1:05 PM
The guidelines "aim to streamline VEP development, sharing, and evaluation by tackling data availability, interpretability, transparency, and circularity." Benjamin J. Livesey, @jmarshlab.bsky.social et al
genomebiology.biomedcentral.com/articles/10....
genomebiology.biomedcentral.com/articles/10....
Following our variant effect predictor (VEP) guidelines paper last week, we’re excited to announce another publication in Genome Biology today—the latest iteration of our VEP benchmarking efforts.
With so many VEPs released recently, how do we choose the best ones?
🌐 doi.org/10.1186/s130...
With so many VEPs released recently, how do we choose the best ones?
🌐 doi.org/10.1186/s130...

Variant effect predictor correlation with functional assays is reflective of clinical classification performance - Genome Biology
Background Understanding the relationship between protein sequence and function is crucial for accurate classification of missense variants. Variant effect predictors (VEPs) play a vital role in decip...
doi.org
April 22, 2025 at 4:02 PM
Following our variant effect predictor (VEP) guidelines paper last week, we’re excited to announce another publication in Genome Biology today—the latest iteration of our VEP benchmarking efforts.
With so many VEPs released recently, how do we choose the best ones?
🌐 doi.org/10.1186/s130...
With so many VEPs released recently, how do we choose the best ones?
🌐 doi.org/10.1186/s130...
New paper out in Genome Biology! 🎉
We lay out best-practice guidelines for releasing variant effect predictors, developed through the Atlas of Variant Effects Alliance @varianteffect.bsky.social
Open, interpretable, and clinically useful VEPs are the goal.
📄 doi.org/10.1186/s130...
We lay out best-practice guidelines for releasing variant effect predictors, developed through the Atlas of Variant Effects Alliance @varianteffect.bsky.social
Open, interpretable, and clinically useful VEPs are the goal.
📄 doi.org/10.1186/s130...

Guidelines for releasing a variant effect predictor - Genome Biology
Computational methods for assessing the likely impacts of mutations, known as variant effect predictors (VEPs), are widely used in the assessment and interpretation of human genetic variation, as well...
doi.org
April 15, 2025 at 12:24 PM
New paper out in Genome Biology! 🎉
We lay out best-practice guidelines for releasing variant effect predictors, developed through the Atlas of Variant Effects Alliance @varianteffect.bsky.social
Open, interpretable, and clinically useful VEPs are the goal.
📄 doi.org/10.1186/s130...
We lay out best-practice guidelines for releasing variant effect predictors, developed through the Atlas of Variant Effects Alliance @varianteffect.bsky.social
Open, interpretable, and clinically useful VEPs are the goal.
📄 doi.org/10.1186/s130...
Great to see you Sarah!
With TeichLab alumni Dr Jing Su and Prof Joe Marsh in lovely Edinburgh at "hidden cell, dark genome conference"

April 4, 2025 at 5:52 PM
Great to see you Sarah!
Reposted by Joe Marsh
Structure-informed classification of RyR1 variants highlights limitations of current predictors and enables clinical interpretation https://www.medrxiv.org/content/10.1101/2025.04.02.25325085v1
April 3, 2025 at 1:40 PM
Structure-informed classification of RyR1 variants highlights limitations of current predictors and enables clinical interpretation https://www.medrxiv.org/content/10.1101/2025.04.02.25325085v1
Had a good time discussing variant effect predictors on this podcast, thanks for having me!
🆕 🎧 Podcast Episode “Deep Thought”
We can predict the weather, but can we predict genetic diseases from your genome? Learn about variant effect predictors (VEPs) with experts Drs Debbie Marks and Joe Marsh
#Podcast #Science #AI #VariantEffectPredictor
🎙 www.varianteffect.org/podcast
We can predict the weather, but can we predict genetic diseases from your genome? Learn about variant effect predictors (VEPs) with experts Drs Debbie Marks and Joe Marsh
#Podcast #Science #AI #VariantEffectPredictor
🎙 www.varianteffect.org/podcast

April 1, 2025 at 4:10 PM
Had a good time discussing variant effect predictors on this podcast, thanks for having me!
Reposted by Joe Marsh
Sign up now for the 'Enter the Dark Genome - Instructions Hidden in Plain Sight' talk by @katarney.bsky.social at @rcpedin.bsky.social on 2 April from 6-7pm, followed by a drinks reception: edin.ac/4ip4egz
March 20, 2025 at 3:01 PM
Sign up now for the 'Enter the Dark Genome - Instructions Hidden in Plain Sight' talk by @katarney.bsky.social at @rcpedin.bsky.social on 2 April from 6-7pm, followed by a drinks reception: edin.ac/4ip4egz
Reposted by Joe Marsh
📣 We are advertising for a postdoc to join our team at the University of Edinburgh! Our lab studies gene regulatory mechanisms in development, and how genetic changes may impact these processes to alter development and shape human craniofacial form and function 🧬🧪
Postdoctoral Researcher
Our research is focused on understanding how genetic changes in the non-coding genome can impact gene regulatory mechanisms, alter developmental processes and ultimately affect human craniofacial shap...
elxw.fa.em3.oraclecloud.com
March 20, 2025 at 9:22 AM
📣 We are advertising for a postdoc to join our team at the University of Edinburgh! Our lab studies gene regulatory mechanisms in development, and how genetic changes may impact these processes to alter development and shape human craniofacial form and function 🧬🧪
Check out our new preprint and Google Colab notebook if you are interested in predicting the molecular mechanisms of missense disease phenotypes. We find that gain-of-function and dominant-negative effects are very common, and that many disease genes are associated with multiple distinct mechanisms
1/ Excited to share our latest work on the "Prevalence of loss-of-function, gain-of-function and dominant-negative mechanisms across genetic disease phenotypes". @marshlab.bsky.social @uoe-igc.bsky.social www.biorxiv.org/content/10.1...

Prevalence of loss-of-function, gain-of-function and dominant-negative mechanisms across genetic disease phenotypes
Molecular disease mechanisms caused by mutations in protein-coding regions are diverse, but they can be broadly categorised into loss-of-function (LOF), gain-of-function (GOF), and dominant-negative (...
www.biorxiv.org
March 17, 2025 at 1:40 PM
Check out our new preprint and Google Colab notebook if you are interested in predicting the molecular mechanisms of missense disease phenotypes. We find that gain-of-function and dominant-negative effects are very common, and that many disease genes are associated with multiple distinct mechanisms


