Where to go after AlphaFold? How do we avoid the field becoming a load of half-baked LLMs?
Let me know what you think.
jgreener64.github.io/posts/struct...
pubs.aip.org/aip/jcp/arti...
pubs.aip.org/aip/jcp/arti...
Do try it out if you like flexible molecular simulations. More features and performance coming soon.
github.com/JuliaMolSim/...
Do try it out if you like flexible molecular simulations. More features and performance coming soon.
github.com/JuliaMolSim/...
Apply at achira.ai

Apply at achira.ai
www.biorxiv.org/content/10.1...
#teamtomo

www.biorxiv.org/content/10.1...
#teamtomo
Where to go after AlphaFold? How do we avoid the field becoming a load of half-baked LLMs?
Let me know what you think.
jgreener64.github.io/posts/struct...
Where to go after AlphaFold? How do we avoid the field becoming a load of half-baked LLMs?
Let me know what you think.
jgreener64.github.io/posts/struct...
www.open-bio.org/2025/09/30/2...

www.open-bio.org/2025/09/30/2...
www.experimental-history.com/p/thank-you-...

www.experimental-history.com/p/thank-you-...
Having people in the same space talking over a long period of time (months) is one way to address this. It tends to happen organically, though, and is hard to plan.
Having people in the same space talking over a long period of time (months) is one way to address this. It tends to happen organically, though, and is hard to plan.
This one explores the looming peer review crisis. As many of you know, it's becoming significantly more difficult for journal editors to find scholars willing to serve as peer reviewers for submitted manuscripts.

This one explores the looming peer review crisis. As many of you know, it's becoming significantly more difficult for journal editors to find scholars willing to serve as peer reviewers for submitted manuscripts.
📄 www.biorxiv.org/content/10.1...
🌐 search.foldseek.com/folddisco
📄 www.biorxiv.org/content/10.1...
🌐 search.foldseek.com/folddisco
www.science.org/content/arti...

www.science.org/content/arti...
You’ll help develop novel molecular simulation methods to optimise sequences for diverse design tasks in computational protein design.
See: www.nature.com/naturecareer...
Apply by 13 JUL
#PostdocJobs #ScienceJobs

You’ll help develop novel molecular simulation methods to optimise sequences for diverse design tasks in computational protein design.
See: www.nature.com/naturecareer...
Apply by 13 JUL
#PostdocJobs #ScienceJobs
www.nature.com/naturecareer...
Feel free to message me with any questions.

www.nature.com/naturecareer...
Feel free to message me with any questions.
YouTube: www.youtube.com/watch?v=NZlE...
Apple: podcasts.apple.com/us/podcast/m...
Spotify: open.spotify.com/episode/1suM...

YouTube: www.youtube.com/watch?v=NZlE...
Apple: podcasts.apple.com/us/podcast/m...
Spotify: open.spotify.com/episode/1suM...
🛠️ Develop #JuliaLang & Python tools to model structural and evolutionary features of IDPs
📅 Start: 1 Sept 2025
🎓 3+ years of higher education (Master’s or engineering diploma preferred)
👉 Apply now: emploi.cnrs.fr/Offres/CDD/U...
🛠️ Develop #JuliaLang & Python tools to model structural and evolutionary features of IDPs
📅 Start: 1 Sept 2025
🎓 3+ years of higher education (Master’s or engineering diploma preferred)
👉 Apply now: emploi.cnrs.fr/Offres/CDD/U...
doi.org/10.26434/che...

doi.org/10.26434/che...
www.nature.com/articles/s41...
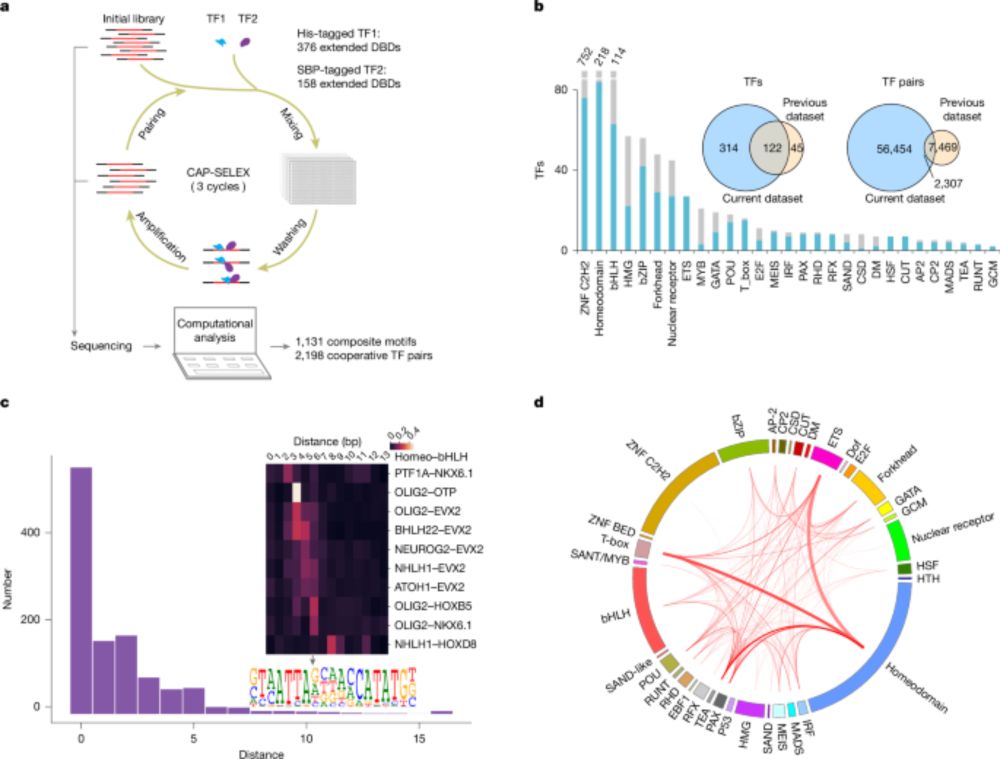
www.nature.com/articles/s41...
Now including an example of fine-tuning MACE-OFF23 to fit the RDF of water. Thanks to the reviewers for useful comments.
Now including an example of fine-tuning MACE-OFF23 to fit the RDF of water. Thanks to the reviewers for useful comments.
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
pubs.acs.org/doi/10.1021/...

These methods are gradually making MD an important part of the drug discovery pipeline.
#compChem
These methods are gradually making MD an important part of the drug discovery pipeline.



