timePlex (multiplexing with time-offsets): www.biorxiv.org/content/10.1...
JMod (search software for joint-modeling): www.biorxiv.org/content/10.1...
PSMtag (novel mass tag for 9-plexDIA): www.biorxiv.org/content/10.1...
timePlex (multiplexing with time-offsets): www.biorxiv.org/content/10.1...
JMod (search software for joint-modeling): www.biorxiv.org/content/10.1...
PSMtag (novel mass tag for 9-plexDIA): www.biorxiv.org/content/10.1...
This enables 27-plex, and we achieve >500 SPD with 25 minutes active chromatography per sample!
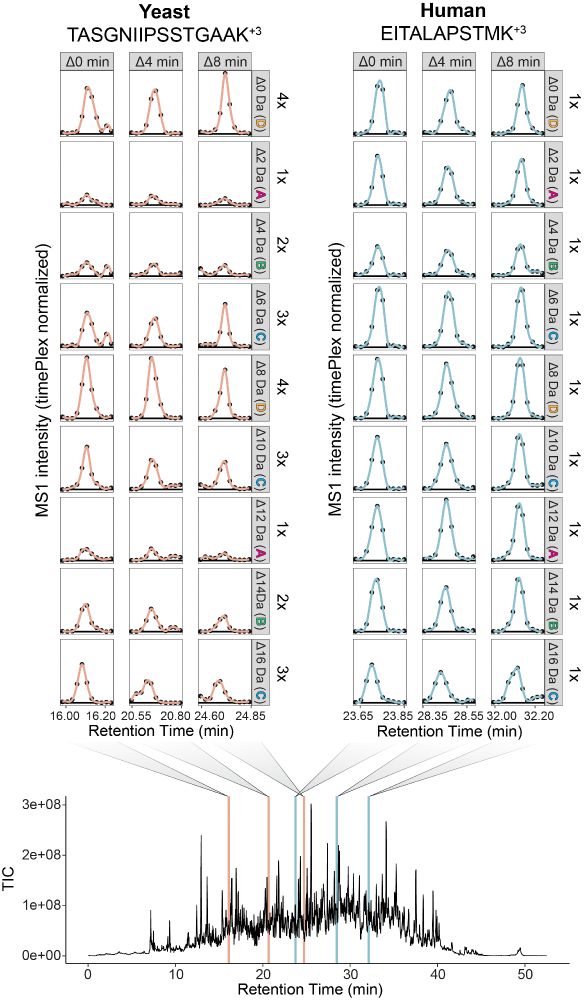
This enables 27-plex, and we achieve >500 SPD with 25 minutes active chromatography per sample!
We stagger and overlap the chromatography of multiple samples in the same run. The time-offsets between samples encode sample-specificity.

We stagger and overlap the chromatography of multiple samples in the same run. The time-offsets between samples encode sample-specificity.
Time and mass dimensions are orthogonal, which unlocks combinatorial multiplexing when timePlex and plexDIA are used together! This introduces the potential for quadratically increased throughput!

Time and mass dimensions are orthogonal, which unlocks combinatorial multiplexing when timePlex and plexDIA are used together! This introduces the potential for quadratically increased throughput!

