https://jsilve24.github.io/SilvermanLab/
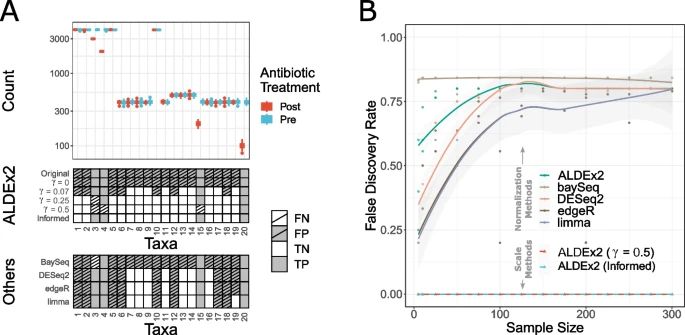
Core takeaway, its important to accurately model uncertainty and error.
@ggloor.bsky.social
Core takeaway, its important to accurately model uncertainty and error.
@ggloor.bsky.social
arxiv.org/abs/2201.03616

arxiv.org/abs/2201.03616
github.com/jsilve24/ALD...
github.com/jsilve24/ALD...
jmlr.org/papers/v23/1...
where we introduced the CU Sampler for Bayesian MLN models. This is even 1-2 orders of magnitude faster than those methods while still be extreemly accurate.
jmlr.org/papers/v23/1...
where we introduced the CU Sampler for Bayesian MLN models. This is even 1-2 orders of magnitude faster than those methods while still be extreemly accurate.

