https://jsilve24.github.io/SilvermanLab/
We have developed specialized PIMs that account for uncertainty in sparsity assumptions. 6 datasets with ground truth, comparing against 8 methods. When our assumptions hold (first 4 datasets) our methods do well. When violated (last two) they fail gracefully.

We have developed specialized PIMs that account for uncertainty in sparsity assumptions. 6 datasets with ground truth, comparing against 8 methods. When our assumptions hold (first 4 datasets) our methods do well. When violated (last two) they fail gracefully.

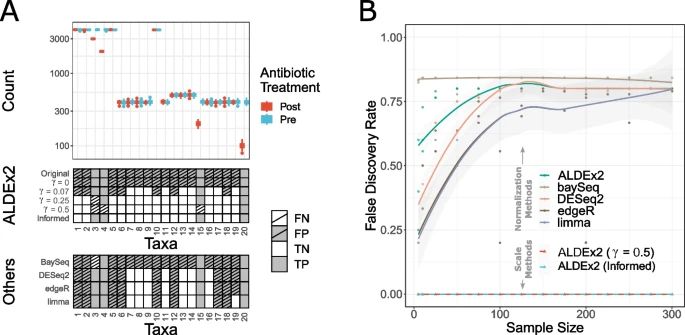
3 real datasets (1 more coming in revision), compared against 5 competing methods (ALDEx2, BayeSeq, DESeq2, edgeR, limma).

3 real datasets (1 more coming in revision), compared against 5 competing methods (ALDEx2, BayeSeq, DESeq2, edgeR, limma).
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
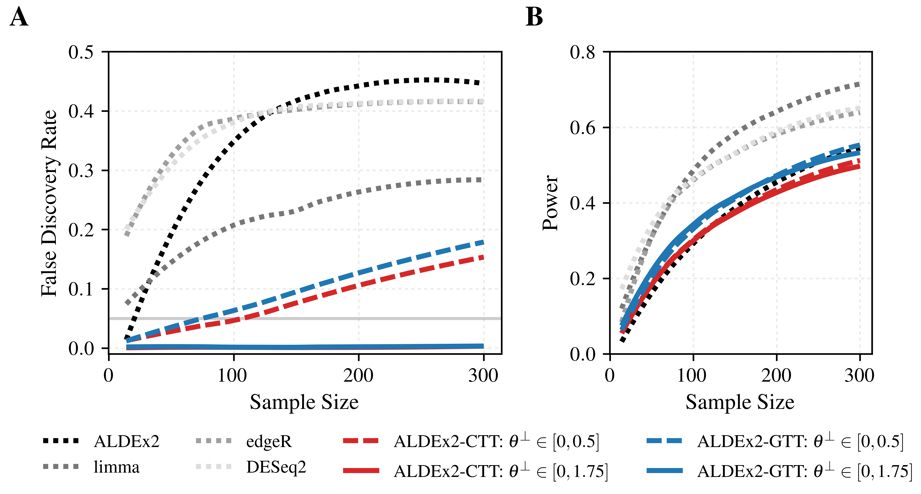
Even slight errors in normalization assumptions about scale (epsilon not equal to zero in this plot) can invalidate conclusions drawn from current methods.

Even slight errors in normalization assumptions about scale (epsilon not equal to zero in this plot) can invalidate conclusions drawn from current methods.
From www.biorxiv.org/content/10.1... (in review at Genome Biology).

From www.biorxiv.org/content/10.1... (in review at Genome Biology).

