https://jsilve24.github.io/SilvermanLab/
We submitted a presubmission inquiry on 9/12 and followed up again on 9/24. We have not heard a response. Is this typical? Could you please help us, we are trying to confirm how we should submit, as a matters arising or as a research article
www.biorxiv.org/content/10.1...

We submitted a presubmission inquiry on 9/12 and followed up again on 9/24. We have not heard a response. Is this typical? Could you please help us, we are trying to confirm how we should submit, as a matters arising or as a research article
www.biorxiv.org/content/10.1...
Machine learning models that attempt to predict microbial load collapse outside of their training context with an R2<0!
In contrast, our Bayesian Partially Identified Models embrace uncertainty in unmeasured microbial load and consistently outpreform.
www.biorxiv.org/content/10.1...

Machine learning models that attempt to predict microbial load collapse outside of their training context with an R2<0!
In contrast, our Bayesian Partially Identified Models embrace uncertainty in unmeasured microbial load and consistently outpreform.
www.biorxiv.org/content/10.1...
PCR bias doesn’t just distort relative abundances—it reshapes microbiome ecological analyses.
We show that commonly used diversity metrics (e.g., UniFrac or Shannon) are not robust to amplification bias, while perturbation-invariant alternatives are.
www.biorxiv.org/content/10.1...

PCR bias doesn’t just distort relative abundances—it reshapes microbiome ecological analyses.
We show that commonly used diversity metrics (e.g., UniFrac or Shannon) are not robust to amplification bias, while perturbation-invariant alternatives are.
www.biorxiv.org/content/10.1...
We relax normalizations to produce statistical methods for bioinformatics that are much more robust and powerful. We see FDR drop from 45% to 5% with increases in power!
This adds to our ongoing work on Scale Reliant Inference.
link.springer.com/article/10.1...

We relax normalizations to produce statistical methods for bioinformatics that are much more robust and powerful. We see FDR drop from 45% to 5% with increases in power!
This adds to our ongoing work on Scale Reliant Inference.
link.springer.com/article/10.1...
Thread explaining the key points below.
journals.asm.org/doi/10.1128/...
Thread explaining the key points below.
journals.asm.org/doi/10.1128/...
genomebiology.biomedcentral.com/articles/10....
We introduce scale models, a generalization of normalizations that explciitly account for uncertainty in biological system scale (e.g., microbial load).
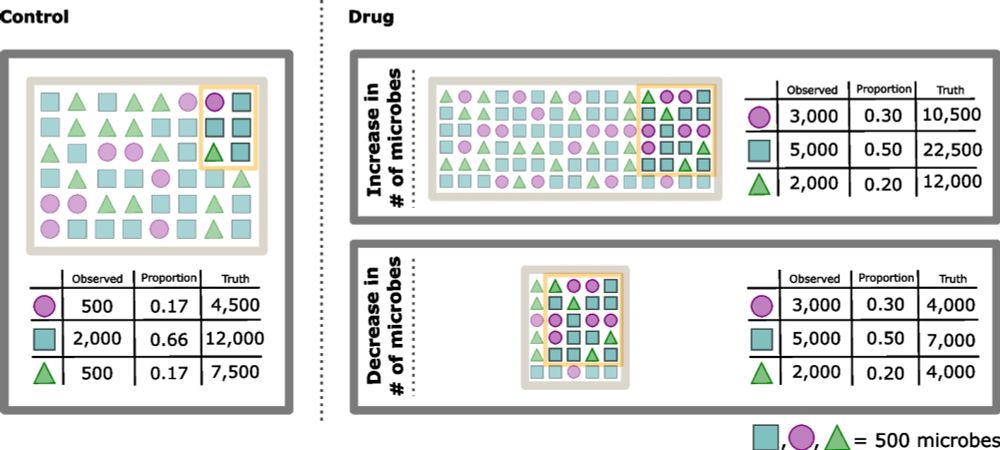
genomebiology.biomedcentral.com/articles/10....
We introduce scale models, a generalization of normalizations that explciitly account for uncertainty in biological system scale (e.g., microbial load).
"Senator Fetterman wants to hear from you about how the federal funding freeze is affecting Pennsylvania."
"If your project has been impacted, please fill out our constituent impact form:" forms.office.com/g/mFv2JAPxpC
Get out your Other Support and share that info!
"Senator Fetterman wants to hear from you about how the federal funding freeze is affecting Pennsylvania."
"If your project has been impacted, please fill out our constituent impact form:" forms.office.com/g/mFv2JAPxpC
Get out your Other Support and share that info!

neurips.cc/virtual/2024...
Also includes extreemly fast marginal likelihood estimation for hyperparameter tuning.
cran.r-project.org/web/packages...
neurips.cc/virtual/2024...
Also includes extreemly fast marginal likelihood estimation for hyperparameter tuning.
cran.r-project.org/web/packages...
arxiv.org/abs/2410.05548
Flexible Multinomial Logistic-Normal time series models (state space models) that scale to extreemly large datasets. Inference is 5-6 orders of magnitude faster than alternatives. R package will soon be released.

arxiv.org/abs/2410.05548
Flexible Multinomial Logistic-Normal time series models (state space models) that scale to extreemly large datasets. Inference is 5-6 orders of magnitude faster than alternatives. R package will soon be released.
We know, we have better methods that have been validated against datasets with ground truth.
www.biorxiv.org/content/10.1...
(thread)

We know, we have better methods that have been validated against datasets with ground truth.
www.biorxiv.org/content/10.1...
(thread)
I would love to join the starter pack -- I am just transitioning form twitter.
Thanks!
I would love to join the starter pack -- I am just transitioning form twitter.
Thanks!

