
🥝 Previously studying plant pathogen evolution with everyone's favourite P. syringae pathovar - Psa! ✨
🌿 hemara.nz
Questions? Reach out to us (Olivia and @zoejolylopez.bsky.social)

Questions? Reach out to us (Olivia and @zoejolylopez.bsky.social)



academic.oup.com/g3journal/ad...

academic.oup.com/g3journal/ad...
Pls repost 🙏
www.evolbio.mpg.de/3838377/job_...

Pls repost 🙏
www.evolbio.mpg.de/3838377/job_...

www.sciencedirect.com/science/arti...

www.sciencedirect.com/science/arti...
View them on isme-microbes.org/isme20-keyno...
#isme20

View them on isme-microbes.org/isme20-keyno...
#isme20

www.nature.com/articles/s41...
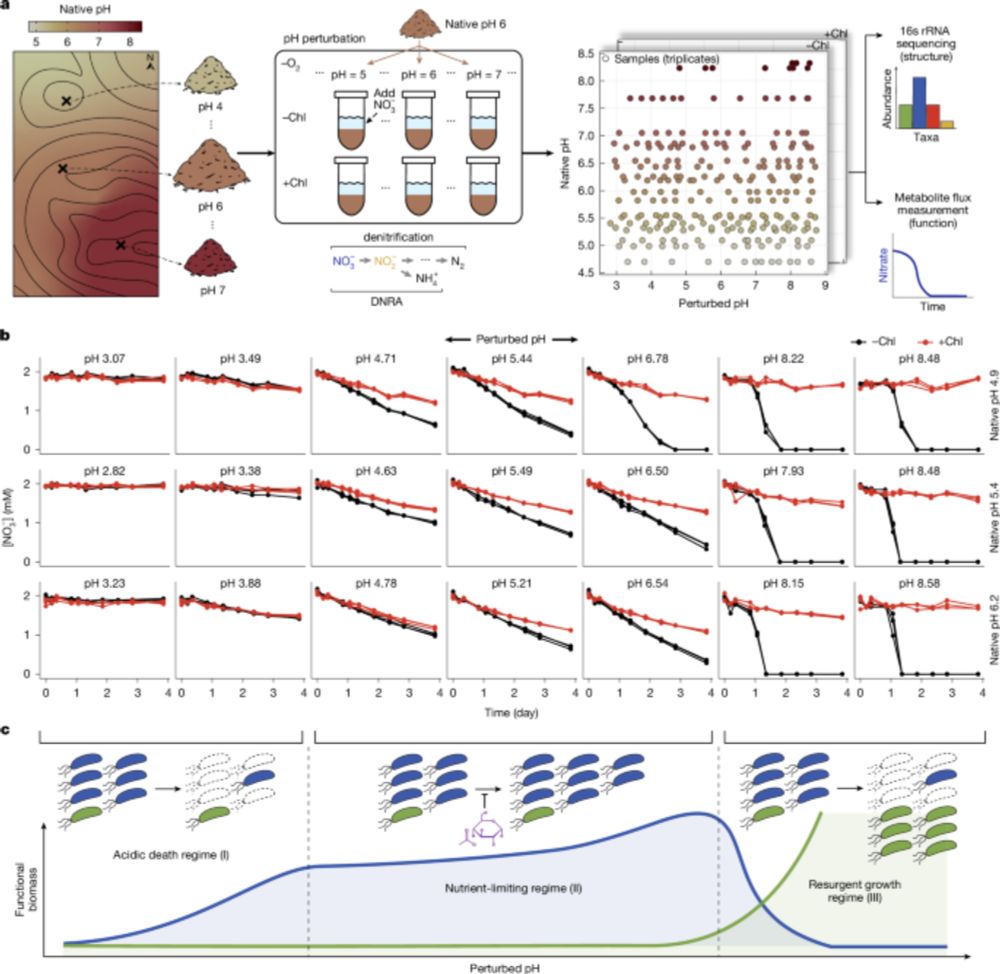
www.nature.com/articles/s41...

We analyzed foliar disease suppressive microbial communities from tomato to identify taxa and functions associated with suppression. Congratulations to lead author @hanareiaehau.bsky.social, and co-authors Javad Sadeghi and Terrence Bell!
tinyurl.com/4pnxxuus

We analyzed foliar disease suppressive microbial communities from tomato to identify taxa and functions associated with suppression. Congratulations to lead author @hanareiaehau.bsky.social, and co-authors Javad Sadeghi and Terrence Bell!
tinyurl.com/4pnxxuus
#MicroSky #PlantScience #Pseudomonas
Our new research in Nature Microbiology uncovers the sophisticated teamwork of Pseudomonas syringae, a notorious plant pathogen.
🔗 rdcu.be/egczU
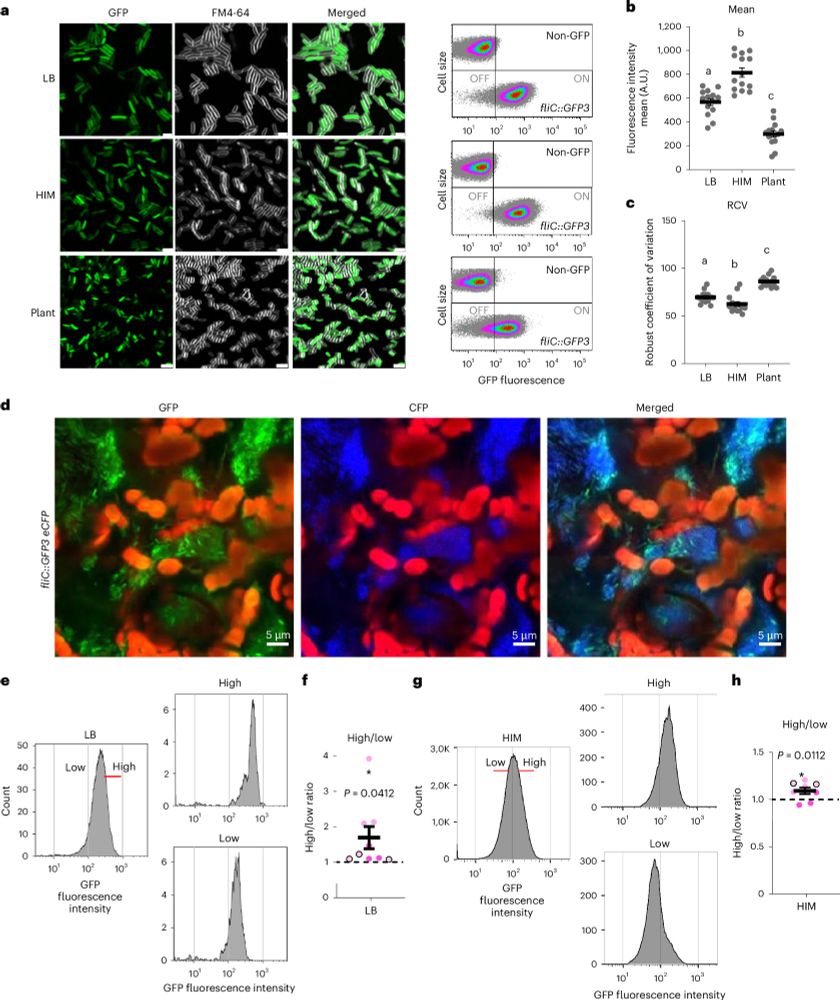
#MicroSky #PlantScience #Pseudomonas
Our new research in Nature Microbiology uncovers the sophisticated teamwork of Pseudomonas syringae, a notorious plant pathogen.
🔗 rdcu.be/egczU
www.nature.com/articles/s41...
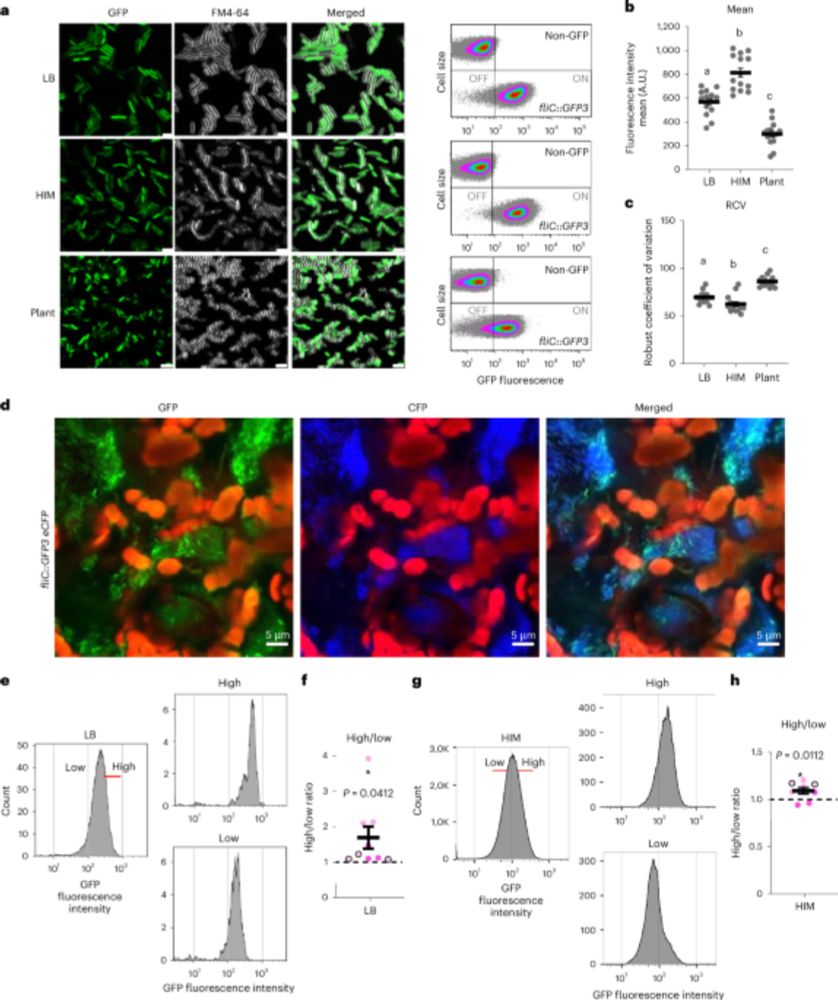
www.nature.com/articles/s41...

enviromicro-journals.onlinelibrary.wiley.com/doi/10.1111/...
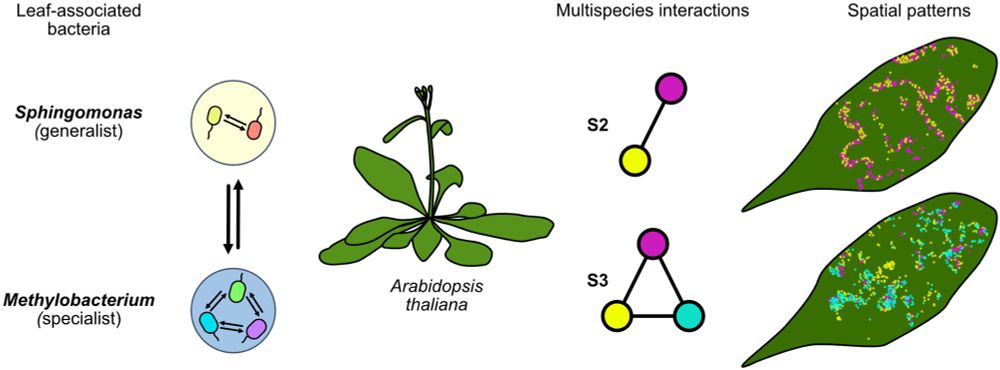
enviromicro-journals.onlinelibrary.wiley.com/doi/10.1111/...
onlinelibrary.wiley.com/doi/abs/10.1...

onlinelibrary.wiley.com/doi/abs/10.1...


We’re hiring a PhD student to start Sept 2025: how do bacteria ‘tame’ phage parasites, and turn them into deadly Tailocins?
Come join me, @brockhurstlab.bsky.social, Patrick Cai &
@raveentank.bsky.social as we study ZOMBIE PHAGES!
findaphd.com/phds/project...
We’re hiring a PhD student to start Sept 2025: how do bacteria ‘tame’ phage parasites, and turn them into deadly Tailocins?
Come join me, @brockhurstlab.bsky.social, Patrick Cai &
@raveentank.bsky.social as we study ZOMBIE PHAGES!
findaphd.com/phds/project...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...


