
🔗 academic.oup.com/gbe
🏠 @official-smbe.bsky.social
🤝 @molbioevol.bsky.social
#genome #evolution #science #biology #societyjournal

🔗 doi.org/10.1093/gbe/evaf196
#genome #evolution
🔗 doi.org/10.1093/gbe/evaf213
#genome #evolution #evodevo

🔗 doi.org/10.1093/gbe/evaf213
#genome #evolution #evodevo
🔗 doi.org/10.1093/gbe/evaf212
#genome #evolution

🔗 doi.org/10.1093/gbe/evaf212
#genome #evolution
🔗 doi.org/10.1093/gbe/evaf200
#genome #evolution #phylogenetics

🔗 doi.org/10.1093/gbe/evaf200
#genome #evolution #phylogenetics
🔗 doi.org/10.1093/gbe/evaf206
#genome #evolution #aDNA #denisova

🔗 doi.org/10.1093/gbe/evaf206
#genome #evolution #aDNA #denisova
🔗 doi.org/10.1093/gbe/evaf207
#genome #evolution

🔗 doi.org/10.1093/gbe/evaf207
#genome #evolution
Read paper in Genome Biology and Evolution: academic.oup.com/gbe/advance-...

Read paper in Genome Biology and Evolution: academic.oup.com/gbe/advance-...
The male-killing gene wmk in Wolbachia splits into 5 types with distinct genomic patterns. Variation in copy number and expression helps explain male-biased lethality across hosts.
Read more: academic.oup.com/gbe/article/...

The male-killing gene wmk in Wolbachia splits into 5 types with distinct genomic patterns. Variation in copy number and expression helps explain male-biased lethality across hosts.
Read more: academic.oup.com/gbe/article/...
🔗 doi.org/10.1093/gbe/evaf208
#genome #evolution

🔗 doi.org/10.1093/gbe/evaf208
#genome #evolution
🔗 doi.org/10.1093/gbe/evaf188
#genome #evolution

🔗 doi.org/10.1093/gbe/evaf188
#genome #evolution
Main findings in the 🧵 below ⬇️
@cpgsthlm.bsky.social @genomebiolevol.bsky.social #aDNA
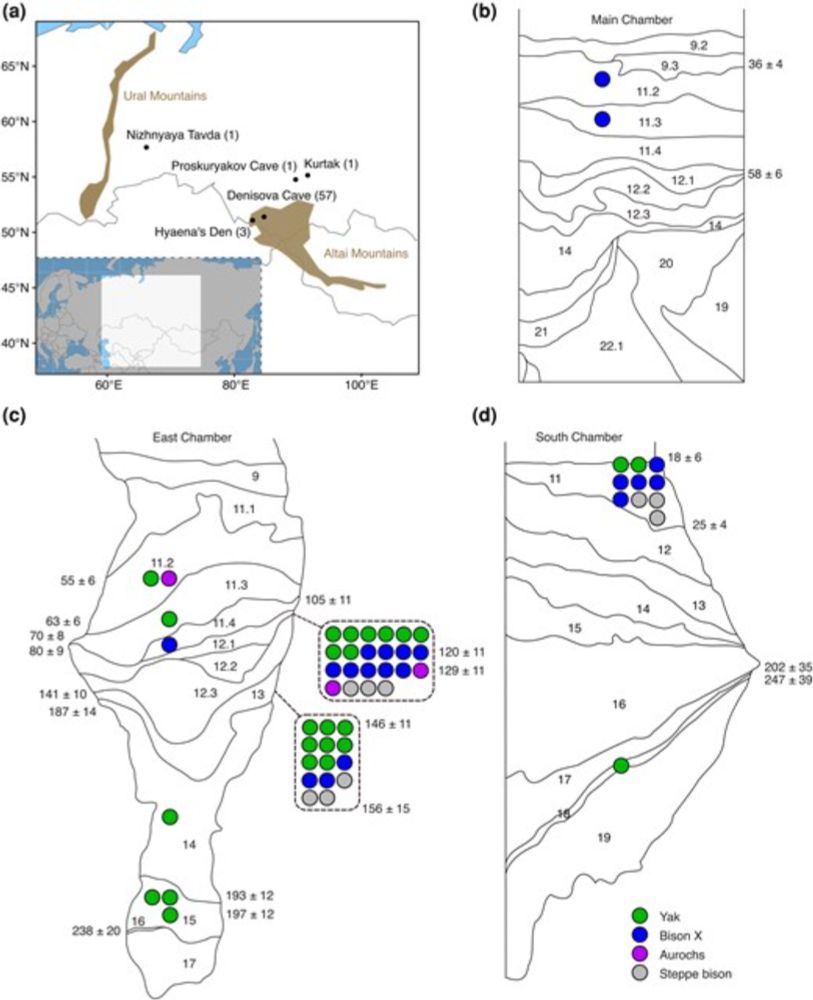
Main findings in the 🧵 below ⬇️
@cpgsthlm.bsky.social @genomebiolevol.bsky.social #aDNA
🔗 doi.org/10.1093/gbe/evaf205
#genome #evolution #birds

🔗 doi.org/10.1093/gbe/evaf205
#genome #evolution #birds
🔗 doi.org/10.1093/gbe/evaf197
#genome #evolution

🔗 doi.org/10.1093/gbe/evaf197
#genome #evolution
🔗 doi.org/10.1093/gbe/evaf199
#genome #evolution #snail

🔗 doi.org/10.1093/gbe/evaf199
#genome #evolution #snail

🔗 doi.org/10.1093/gbe/evaf199
#genome #evolution #snail
🔗 doi.org/10.1093/gbe/evaf181
#genome #evolution #domestication

🔗 doi.org/10.1093/gbe/evaf181
#genome #evolution #domestication


🔗 doi.org/10.1093/gbe/evaf201
#genome #evolution #CRISPR

🔗 doi.org/10.1093/gbe/evaf201
#genome #evolution #CRISPR
Precious collaboration w/ colleagues across the world, led alongside the amazing @mbrasovives.bsky.social and the one&only @jrotwitguez.bsky.social
Check out his great thread!
Precious collaboration w/ colleagues across the world, led alongside the amazing @mbrasovives.bsky.social and the one&only @jrotwitguez.bsky.social
Check out his great thread!
Have a read!! ➡️ doi.org/10.1093/gbe/...
@emilruff.bsky.social @alexjaffe.bsky.social @geomicrosoares.bsky.social & others not in Bluesky!

Have a read!! ➡️ doi.org/10.1093/gbe/...
@emilruff.bsky.social @alexjaffe.bsky.social @geomicrosoares.bsky.social & others not in Bluesky!
Our latest perspective piece is out, and it’s one you won’t want to miss- 'The Genomic Kaleidoscope'
academic.oup.com/gbe/article/...
Co-lead by @mbrasovives.bsky.social @jrotwitguez.bsky.social & @diegoharta.bsky.social
📌 Check the full post here👇
Our latest perspective piece is out, and it’s one you won’t want to miss- 'The Genomic Kaleidoscope'
academic.oup.com/gbe/article/...
Co-lead by @mbrasovives.bsky.social @jrotwitguez.bsky.social & @diegoharta.bsky.social
📌 Check the full post here👇
🔗 doi.org/10.1093/gbe/evaf204
#genome #evolution

🔗 doi.org/10.1093/gbe/evaf204
#genome #evolution

🔗 doi.org/10.1093/gbe/evaf204
#genome #evolution

🔗 doi.org/10.1093/gbe/evaf204
#genome #evolution
"The Genomic Kaleidoscope: On the Hidden Dimensions of Within-Species Genomic Diversity" 💡🌀🌈
doi.org/10.1093/gbe/...
Co-led with @mbrasovives.bsky.social and @diegoharta.bsky.social
Check out our thread! 🧵👇 (1/n)

