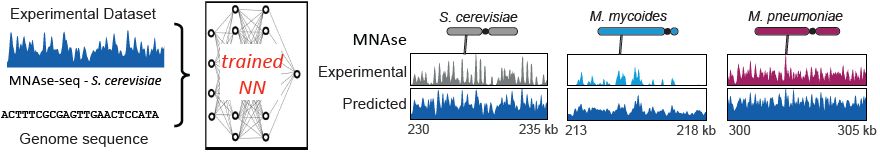
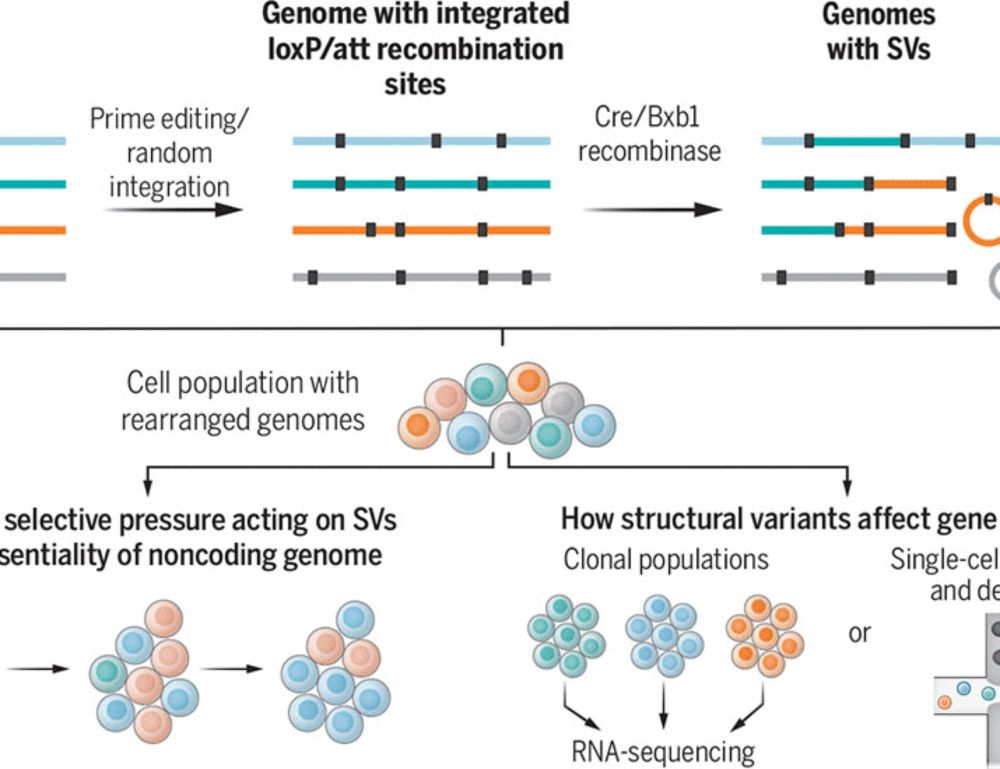
...explained by known motifs of its own (increased overall affinity) or other TF (thus representing TF-TF interaction/cooperation), *or some other unknown factors*
...explained by known motifs of its own (increased overall affinity) or other TF (thus representing TF-TF interaction/cooperation), *or some other unknown factors*