
Daniel Friedrich
@friedrichlab.bsky.social
Research Group Leader @chemunicologne.bsky.social at University of Cologne (@unicologne.bsky.social) in Biomolecular #NMR Spectroscopy | We are interested in structural analysis of proteins, peptides and nucleic acids.
https://nmr.chemie.uni-koeln.de/
https://nmr.chemie.uni-koeln.de/
Very nice article from @lek-lab.bsky.social including @roesslph.bsky.social, A. Sever and R. Ahmed, enjoyed reading it! #NMR
Solution NMR goes big: Atomic resolution studies of protein components of molecular machines and phase-separated condensates, Alexander I.M. Sever, Rashik Ahmed, Philip Rößler, Lewis E. Kay* @lek-lab.bsky.social @utoronto.ca www.sciencedirect.com/science/arti... #NMRchat 🧲

Solution NMR goes big: Atomic resolution studies of protein components of molecular machines and phase-separated condensates
The tools of structural biology have undergone remarkable advances in the past decade. These include new computational and experimental approaches tha…
www.sciencedirect.com
November 10, 2025 at 5:53 PM
Very nice article from @lek-lab.bsky.social including @roesslph.bsky.social, A. Sever and R. Ahmed, enjoyed reading it! #NMR
Fantastic opportunity to work on exciting projects with a great team @etzkornlab.bsky.social!
We are hiring!
The Etzkorn Lab is looking for up to 7 highly motivated PostDocs and 1-2 PhD candidates to support our endeavour to translate the DNAzyme technology into therapeutic applications. For more information see: www.ipb.hhu.de/fileadmin/re...
Please repost. #Postdoc, @hhu.de
The Etzkorn Lab is looking for up to 7 highly motivated PostDocs and 1-2 PhD candidates to support our endeavour to translate the DNAzyme technology into therapeutic applications. For more information see: www.ipb.hhu.de/fileadmin/re...
Please repost. #Postdoc, @hhu.de

November 5, 2025 at 3:55 PM
Fantastic opportunity to work on exciting projects with a great team @etzkornlab.bsky.social!
Reposted by Daniel Friedrich
Our work on DNA G4 interactions with Zuo1 in collaboration with @paeschkelab.bsky.social and the Penedo lab is out in @narjournal.bsky.social 😍
Full text: academic.oup.com/nar/article/...
#G4 #G-Quadruplex #NMR #smFRET #ChIP-qPCR
Full text: academic.oup.com/nar/article/...
#G4 #G-Quadruplex #NMR #smFRET #ChIP-qPCR

The Zuo1 C-terminal domain stabilizes DNA guanosine quadruplex (G4) structures located on Chromosome IX in Saccharomyces cerevisiae
Abstract. Deoxyguanosine quadruplexes (G4s) form stable non-B-DNA structures that can affect transcription, replication, and genome stability. Depending on
academic.oup.com
November 3, 2025 at 5:18 PM
Our work on DNA G4 interactions with Zuo1 in collaboration with @paeschkelab.bsky.social and the Penedo lab is out in @narjournal.bsky.social 😍
Full text: academic.oup.com/nar/article/...
#G4 #G-Quadruplex #NMR #smFRET #ChIP-qPCR
Full text: academic.oup.com/nar/article/...
#G4 #G-Quadruplex #NMR #smFRET #ChIP-qPCR
Reposted by Daniel Friedrich
📢 Open Call! The Max Perutz Labs invite applications for a Full Professorship in Integrative Structure Biology with a focus on in situ structural biology using cryo-electron tomography (cryo-ET) and related methods. More details ➡️ tinyurl.com/brswbymu

October 31, 2025 at 9:24 AM
📢 Open Call! The Max Perutz Labs invite applications for a Full Professorship in Integrative Structure Biology with a focus on in situ structural biology using cryo-electron tomography (cryo-ET) and related methods. More details ➡️ tinyurl.com/brswbymu
Reposted by Daniel Friedrich
We have progressed from data collection to data analysis.

November 1, 2025 at 12:31 AM
We have progressed from data collection to data analysis.
Reposted by Daniel Friedrich
Darja combines organic-chemistry synthesis (with our great collaborator Roman Lichtenecker) of specifically isotope-labeled amino acids with protein NMR in solution and solids.
👉 Synthesis and proof of principle is here: chemistry-europe.onlinelibrary.wiley.com/doi/full/10....
2/n
👉 Synthesis and proof of principle is here: chemistry-europe.onlinelibrary.wiley.com/doi/full/10....
2/n

Synthesis of Selectively 13C/2H/15N‐ Labeled Arginine to Probe Protein Conformation and Interaction by NMR Spectroscopy
We offer a new labeling approach to introduce isolated 15N−13C−1H spin systems to the side chains of arginine residues. The labeling scheme delivers high-resolution NMR spectra to study large protein....
chemistry-europe.onlinelibrary.wiley.com
October 8, 2025 at 6:12 PM
Darja combines organic-chemistry synthesis (with our great collaborator Roman Lichtenecker) of specifically isotope-labeled amino acids with protein NMR in solution and solids.
👉 Synthesis and proof of principle is here: chemistry-europe.onlinelibrary.wiley.com/doi/full/10....
2/n
👉 Synthesis and proof of principle is here: chemistry-europe.onlinelibrary.wiley.com/doi/full/10....
2/n
Reposted by Daniel Friedrich
Really interesting to see some basic science research coming out of Apple Inc. SimpleFold: Folding Proteins is Simpler than You Think arxiv.org/abs/2509.184...
tldr: Apple used flow matching models to more efficiently predict protein folding structures.
More of this, please... from ALL big tech.
tldr: Apple used flow matching models to more efficiently predict protein folding structures.
More of this, please... from ALL big tech.

SimpleFold: Folding Proteins is Simpler than You Think
Protein folding models have achieved groundbreaking results typically via a combination of integrating domain knowledge into the architectural blocks and training pipelines. Nonetheless, given the suc...
arxiv.org
September 25, 2025 at 1:14 AM
Really interesting to see some basic science research coming out of Apple Inc. SimpleFold: Folding Proteins is Simpler than You Think arxiv.org/abs/2509.184...
tldr: Apple used flow matching models to more efficiently predict protein folding structures.
More of this, please... from ALL big tech.
tldr: Apple used flow matching models to more efficiently predict protein folding structures.
More of this, please... from ALL big tech.
Congratulations to the Behrmann and Schwarz labs at Institute of Biochemistry @chemunicologne.bsky.social for this fantastic work! 🎉 #StructuralBiology in Cologne is on fire 💥
🧠 How Synapses Hold Together 🤝
This #discovery was made by a research team @bccologne.bsky.social studying the molecular architecture of #synapses. The study shows that the protein #gephrin forms flexible filaments in the brain, serving as an important building block of inhibitory synapses.
This #discovery was made by a research team @bccologne.bsky.social studying the molecular architecture of #synapses. The study shows that the protein #gephrin forms flexible filaments in the brain, serving as an important building block of inhibitory synapses.
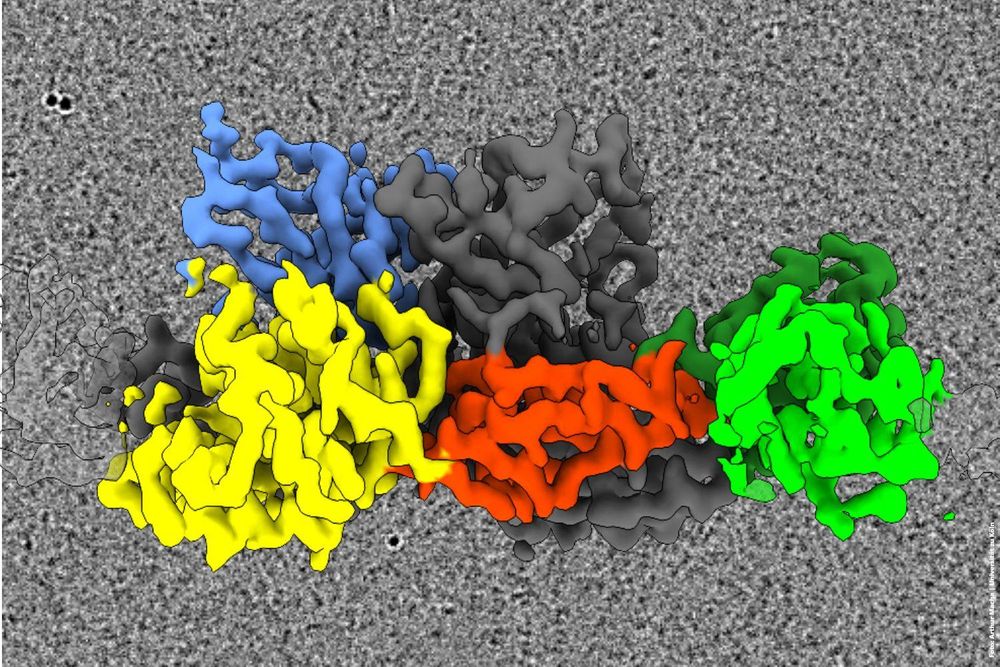
September 24, 2025 at 7:47 PM
Congratulations to the Behrmann and Schwarz labs at Institute of Biochemistry @chemunicologne.bsky.social for this fantastic work! 🎉 #StructuralBiology in Cologne is on fire 💥
I enjoyed the talk by Harald Schwalbe @schwalbe-lab.bsky.social very much, especially the #NMR work on #RNA structure and dynamics! #FGMR #GDCh
At the 2025 meeting of the German focus group of Magnetic Resonance (FGMR, EPR/NMR), Harald Schwalbe, Instruct-director, introduced access possibilities and highlighted the recent installation of the German Instruct centre.
@instruct-eric.bsky.social @gdch.de
#NMR #EPR #FGMR #GDCh #InstructERIC
@instruct-eric.bsky.social @gdch.de
#NMR #EPR #FGMR #GDCh #InstructERIC


September 16, 2025 at 1:48 PM
I enjoyed the talk by Harald Schwalbe @schwalbe-lab.bsky.social very much, especially the #NMR work on #RNA structure and dynamics! #FGMR #GDCh
Reposted by Daniel Friedrich
Postdoctoral position in viral protein NMR, Prof Anja Böckmann, The protein solid-state NMR laboratory within the Molecular Microbiology and Structural Biochemistry (MMSB) unit in the Lyon Gerland Bioscience Campus mmsb.cnrs.fr/en/
emploi.cnrs.fr/Offres/CDD/U... #NMRjobs #NMRchat 🧲
emploi.cnrs.fr/Offres/CDD/U... #NMRjobs #NMRchat 🧲
Portail Emploi CNRS - Offre d'emploi - Postdoctoral position in viral protein NMR (M/F)
emploi.cnrs.fr
September 12, 2025 at 1:03 PM
Postdoctoral position in viral protein NMR, Prof Anja Böckmann, The protein solid-state NMR laboratory within the Molecular Microbiology and Structural Biochemistry (MMSB) unit in the Lyon Gerland Bioscience Campus mmsb.cnrs.fr/en/
emploi.cnrs.fr/Offres/CDD/U... #NMRjobs #NMRchat 🧲
emploi.cnrs.fr/Offres/CDD/U... #NMRjobs #NMRchat 🧲
Very nice work by the Wöhnert group, determining an intersting RNA structure using #NMR!
Exciting work from Duchardt-Ferner et al. in NAR: high-res. NMR structure of a tobramycin-responsive riboswitch. Shows a novel aminoglycoside-binding RNA motif with unique RNA-RNA contacts, explaining its exceptional switching efficiency in eukaryotic translation.
academic.oup.com/nar/article/...
academic.oup.com/nar/article/...

Structural basis for ligand recognition in the tobramycin riboswitch
Abstract. Recently, a novel tobramycin-responsive riboswitch was developed by a combination of Capture-SELEX and in vivo screening. This riboswitch regulat
academic.oup.com
September 8, 2025 at 7:19 PM
Very nice work by the Wöhnert group, determining an intersting RNA structure using #NMR!
Reposted by Daniel Friedrich
Exciting work from Duchardt-Ferner et al. in NAR: high-res. NMR structure of a tobramycin-responsive riboswitch. Shows a novel aminoglycoside-binding RNA motif with unique RNA-RNA contacts, explaining its exceptional switching efficiency in eukaryotic translation.
academic.oup.com/nar/article/...
academic.oup.com/nar/article/...

Structural basis for ligand recognition in the tobramycin riboswitch
Abstract. Recently, a novel tobramycin-responsive riboswitch was developed by a combination of Capture-SELEX and in vivo screening. This riboswitch regulat
academic.oup.com
September 8, 2025 at 3:14 PM
Exciting work from Duchardt-Ferner et al. in NAR: high-res. NMR structure of a tobramycin-responsive riboswitch. Shows a novel aminoglycoside-binding RNA motif with unique RNA-RNA contacts, explaining its exceptional switching efficiency in eukaryotic translation.
academic.oup.com/nar/article/...
academic.oup.com/nar/article/...
Reposted by Daniel Friedrich
New publication: Arginine dynamics probed by magic-angle spinning NMR with a specific isotope-labeling scheme
Our selective arginine labeling yields well-resolved 1H-detected spectra in solids (and solution). We apply it to study dynamics in crystalline ubiquitin and a >130 kDa large enzyme
1/2
Our selective arginine labeling yields well-resolved 1H-detected spectra in solids (and solution). We apply it to study dynamics in crystalline ubiquitin and a >130 kDa large enzyme
1/2
August 25, 2025 at 5:43 AM
New publication: Arginine dynamics probed by magic-angle spinning NMR with a specific isotope-labeling scheme
Our selective arginine labeling yields well-resolved 1H-detected spectra in solids (and solution). We apply it to study dynamics in crystalline ubiquitin and a >130 kDa large enzyme
1/2
Our selective arginine labeling yields well-resolved 1H-detected spectra in solids (and solution). We apply it to study dynamics in crystalline ubiquitin and a >130 kDa large enzyme
1/2
Very interesting #NMR work on disordered proteins by the lab of @millessigrid.bsky.social at @fmp-berlin.de! Enjoyed reading it, congrats to this study!
I am happy to share our new article on dynamic Eps15 and Dab2 networks in endocytosis studied by #NMR, and forming #LLPS. Research was done at @fmp-berlin.de and at the Institut de Biologie Structurale (IBS).
www.nature.com/articles/s41...
www.nature.com/articles/s41...

Promiscuous and multivalent interactions between Eps15 and partner protein Dab2 generate a complex interaction network - Nature Communications
Eps15, a protein involved in endocytosis initiation, uses small folded EH domains to bind disordered partner proteins and its own disordered tail, thus forming a dynamic network via phase separation t...
www.nature.com
August 21, 2025 at 11:57 AM
Very interesting #NMR work on disordered proteins by the lab of @millessigrid.bsky.social at @fmp-berlin.de! Enjoyed reading it, congrats to this study!
Reposted by Daniel Friedrich
📣 Massively proud of this ⬇️ great study, led by the brilliant @mesny.bsky.social surprisingly uncovering that many pathogen effectors stem from ancient antimicrobials 🤯 #EffectorWisdom #EvoMPMI
📣 NEW PREPRINT 📝
We identified evolutionary origins of many fungal effectors!
We show that fungi secrete lots of antimicrobial proteins, and that some of them were repurposed by plant pathogens for host immune suppression.
www.biorxiv.org/content/10.1...
cc @teamthomma.bsky.social
We identified evolutionary origins of many fungal effectors!
We show that fungi secrete lots of antimicrobial proteins, and that some of them were repurposed by plant pathogens for host immune suppression.
www.biorxiv.org/content/10.1...
cc @teamthomma.bsky.social

Plant-associated fungi co-opt ancient antimicrobials for host manipulation
Evolutionary histories of effector proteins secreted by fungal pathogens to mediate plant colonization remain largely elusive. While most functionally characterized effectors modulate plant immunity, ...
www.biorxiv.org
August 15, 2025 at 7:58 AM
📣 Massively proud of this ⬇️ great study, led by the brilliant @mesny.bsky.social surprisingly uncovering that many pathogen effectors stem from ancient antimicrobials 🤯 #EffectorWisdom #EvoMPMI
I‘m already looking forward to join this very interesting #NMR symposium and to meet friends from the good old times at @fmp-berlin.de! It’s still time to register!
⏰Join us for the NMR Symposium, Nov 26–27, 2025, celebrating our new 1.2 GHz NMR spectrometer. To present a poster, send your abstract to NMR2025@fmp-berlin.de. Registrations are open until September 15th!

August 14, 2025 at 9:27 AM
I‘m already looking forward to join this very interesting #NMR symposium and to meet friends from the good old times at @fmp-berlin.de! It’s still time to register!
Such a beautiful cover for @jacs.acspublications.org designed by @barthvanrossum.bsky.social! Nice #NMR work by the Lange lab at @fmp-berlin.de!
Great cover art by @barthvanrossum.bsky.social for @jacs.acspublications.org: Influenza lifecycle showing M2’s roles, from activation in the low-pH endosome to viral uncoating, RNA release, and budding of new particles. View the article from the Lange lab here: pubs.acs.org/doi/10.1021/...

August 13, 2025 at 2:54 PM
Such a beautiful cover for @jacs.acspublications.org designed by @barthvanrossum.bsky.social! Nice #NMR work by the Lange lab at @fmp-berlin.de!
Great work involving #NMR by @delvallelab.bsky.social in @jacs.acspublications.org on intrastrand side chain stapling of peptides, very interesting read!
pubs.acs.org/doi/10.1021/...
pubs.acs.org/doi/10.1021/...

Intrastrand Peptide Staples That Promote β-Sheet Folding, Self-Assembly, and Amyloid Seeding
Side chain stapling of cysteine (Cys) residues offers convenient entry into constrained peptides with enhanced bioactivity and bioavailability. Despite its widespread application in the constraint of α-helical, PPII, and loop conformations, the stabilization of β-sheet folds via intrastrand side chain Cys stapling remains largely unexplored. Here, we demonstrate that i→i+2 stapling with E-butenyl, butynyl, and m-xylyl linkers significantly enhances the folded population of two distinct β-hairpin model peptides. High-resolution NMR structures reveal that these staples support canonical β-sheet backbone torsions and stabilize cross-strand interactions. Leveraging the maintenance of intact backbone hydrogen-bonding edges, we employed i→i+2 side chain macrocyclization in the design of constrained β-arch peptides derived from the tau protein. We show that intrastrand stapling of a nonaggregation-prone segment promotes self-assembly into β-sheet-like filaments. The resulting filaments also seed the aggregation of endogenous tau in a cell-based assay in a macrocycle- and sequence-dependent manner. These findings establish di-Cys i→i+2 stapling as a versatile and synthetically accessible method to stabilize β-sheet structure and modulate the self-assembly of seed-competent amyloidogenic peptides.
pubs.acs.org
August 11, 2025 at 10:26 AM
Great work involving #NMR by @delvallelab.bsky.social in @jacs.acspublications.org on intrastrand side chain stapling of peptides, very interesting read!
pubs.acs.org/doi/10.1021/...
pubs.acs.org/doi/10.1021/...
The complexity of new functionalities enabled by thousands of non-canonical open reading frames potentially coding for miniproteins is just fascinating! I can see a lot of interesting #NMR studies on structure and function of #peptides
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

High-quality peptide evidence for annotating non-canonical open reading frames as human proteins
A major scientific drive is to characterize the protein-coding genome as it provides the primary basis for the study of human health. But the fundamental question remains: what has been missed in prio...
www.biorxiv.org
August 7, 2025 at 1:48 PM
The complexity of new functionalities enabled by thousands of non-canonical open reading frames potentially coding for miniproteins is just fascinating! I can see a lot of interesting #NMR studies on structure and function of #peptides
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Reposted by Daniel Friedrich
Nature research paper: Diffusing protein binders to intrinsically disordered proteins
go.nature.com/4lSCdzE
go.nature.com/4lSCdzE
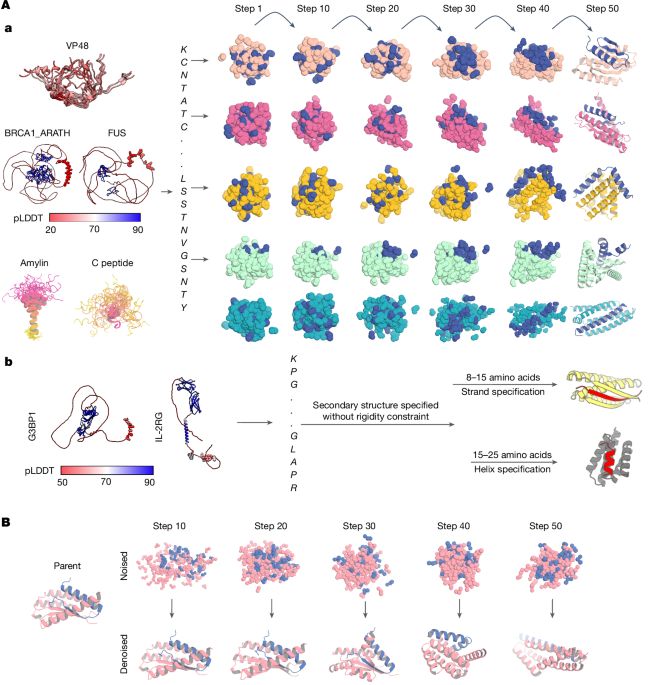
Diffusing protein binders to intrinsically disordered proteins - Nature
Using RFdiffusion, a general method for targeting intrinsically disordered proteins and regions for protein design has been developed.
go.nature.com
July 31, 2025 at 12:58 PM
Nature research paper: Diffusing protein binders to intrinsically disordered proteins
go.nature.com/4lSCdzE
go.nature.com/4lSCdzE
Reposted by Daniel Friedrich
📢Happy to announce that we have been awarded national funding for the ARBICAT project by @ageinves.bsky.social for the next 3 years.
It's a huge honour to be leading this project as a co-PI alongside Pere Clapés, exploring NMR methods for developing new ARtificial BIoCATalysts.
#PID2024 #NMRchat
It's a huge honour to be leading this project as a co-PI alongside Pere Clapés, exploring NMR methods for developing new ARtificial BIoCATalysts.
#PID2024 #NMRchat
July 29, 2025 at 7:52 PM
📢Happy to announce that we have been awarded national funding for the ARBICAT project by @ageinves.bsky.social for the next 3 years.
It's a huge honour to be leading this project as a co-PI alongside Pere Clapés, exploring NMR methods for developing new ARtificial BIoCATalysts.
#PID2024 #NMRchat
It's a huge honour to be leading this project as a co-PI alongside Pere Clapés, exploring NMR methods for developing new ARtificial BIoCATalysts.
#PID2024 #NMRchat
Highly recommended read for everyone interested in interactions of proteins and DNA: Impressive new method on single cell level introduced by @epistrucstab.bsky.social from @unicologne.bsky.social!
🔬 New: Method for mapping protein-DNA interactions developed 💡
@epistrucstab.bsky.social #UniCologne has developed DynaTag, a method that detects the binding of proteins to DNA even in a single cell. This is crucial for gene regulation and the development of diseases like cancer. 🧬👇
uni.koeln/LXJQE
@epistrucstab.bsky.social #UniCologne has developed DynaTag, a method that detects the binding of proteins to DNA even in a single cell. This is crucial for gene regulation and the development of diseases like cancer. 🧬👇
uni.koeln/LXJQE
New method developed for mapping protein binding to DNA
A research team at the University of Cologne has developed a method called DynaTag, which can detect the binding of proteins to DNA, even in an individual cell. Interactions of proteins with DNA are c...
uni.koeln
July 28, 2025 at 10:41 AM
Highly recommended read for everyone interested in interactions of proteins and DNA: Impressive new method on single cell level introduced by @epistrucstab.bsky.social from @unicologne.bsky.social!
Great #NMR work on PTMs of histones by @roesslph.bsky.social and colleagues at @lek-lab.bsky.social!
Fresh off the press from the @lek-lab.bsky.social: Modulation of histone tail electrostatic potentials in nucleosome core particles by acetylation and PARylation
#NMRchat
www.pnas.org/doi/10.1073/...
#NMRchat
www.pnas.org/doi/10.1073/...
PNAS
Proceedings of the National Academy of Sciences (PNAS), a peer reviewed journal of the National Academy of Sciences (NAS) - an authoritative source of high-impact, original research that broadly spans...
www.pnas.org
July 22, 2025 at 11:20 AM
Great #NMR work on PTMs of histones by @roesslph.bsky.social and colleagues at @lek-lab.bsky.social!
We are beyond excited to share our new paper on #NMR structures of antimicrobial #peptides in @pubs.acs.org Biochemistry! 1/3
pubs.acs.org/doi/10.1021/...
pubs.acs.org/doi/10.1021/...

α-Helical Structure of Antimicrobial Peptides Enhances Their Activity through Molecular Surface Signatures
The increase in antibacterial resistance is one of the greatest challenges in modern medicine, driving an urgent need to develop new drugs to combat resistant pathogens. Peptides represent a promising...
pubs.acs.org
July 14, 2025 at 6:26 AM
We are beyond excited to share our new paper on #NMR structures of antimicrobial #peptides in @pubs.acs.org Biochemistry! 1/3
pubs.acs.org/doi/10.1021/...
pubs.acs.org/doi/10.1021/...
Great news for all students @unicologne.bsky.social interested in #structuralbiology, in particular @chemunicologne.bsky.social and at the Department of Biology: We are very honoured to receive funding through the 2025 #Fulbright-Cottrell Award!
July 8, 2025 at 9:54 PM
Great news for all students @unicologne.bsky.social interested in #structuralbiology, in particular @chemunicologne.bsky.social and at the Department of Biology: We are very honoured to receive funding through the 2025 #Fulbright-Cottrell Award!

