Evolution of comparative transcriptomics: biological scales, phylogenetic spans, and modeling frameworks
authors.elsevier.com/sd/article/S...
By @mattezambon.bsky.social & @fedemantica.bsky.social, together with @jonnyfrazer.bsky.social & Mafalda Dias.
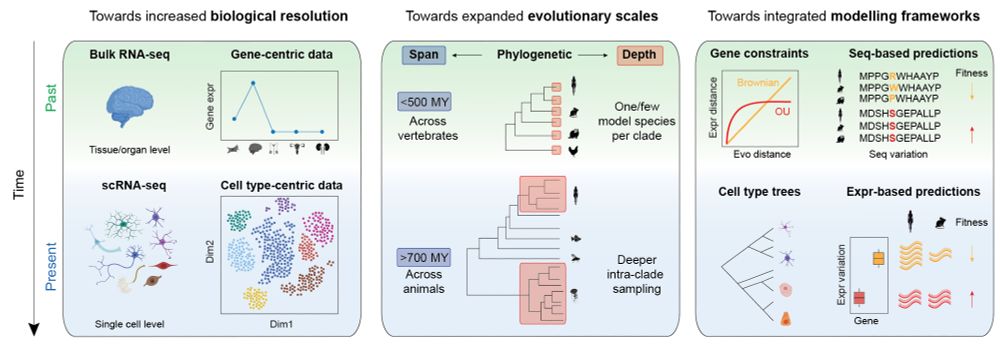
Evolution of comparative transcriptomics: biological scales, phylogenetic spans, and modeling frameworks
authors.elsevier.com/sd/article/S...
By @mattezambon.bsky.social & @fedemantica.bsky.social, together with @jonnyfrazer.bsky.social & Mafalda Dias.
- SIMD FW/BW alignment (preprint soon!)
- Sub. Mat. λ calculator by Eric Dawson
- Faster ARM SW by Alexander Nesterovskiy
- MSA-Pairformer’s proximity-based pairing for multimer prediction (www.biorxiv.org/content/10.1...; avail. in ColabFold API)
💾 github.com/soedinglab/M... & 🐍

- SIMD FW/BW alignment (preprint soon!)
- Sub. Mat. λ calculator by Eric Dawson
- Faster ARM SW by Alexander Nesterovskiy
- MSA-Pairformer’s proximity-based pairing for multimer prediction (www.biorxiv.org/content/10.1...; avail. in ColabFold API)
💾 github.com/soedinglab/M... & 🐍
Stay tuned @workshopmlsb.bsky.social as we share details about the stellar lineup of speakers, the official call for papers, and other announcements!🌟

github.com/martinpacesa...
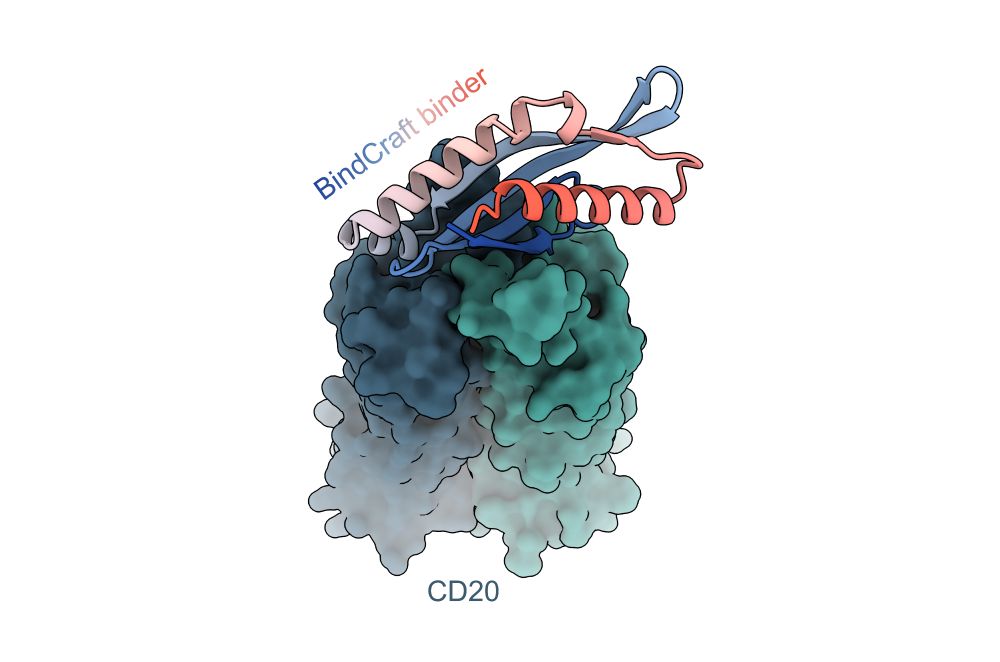
github.com/martinpacesa...
We’ve released a major update to our ProtRL repo:
✅ GRPO via Hugging Face Trainer
✅ New support for weighted DPO
Built for flexible, scalable RL with HF trainer base!
Check here: github.com/AI4PDLab/Pro...

We’ve released a major update to our ProtRL repo:
✅ GRPO via Hugging Face Trainer
✅ New support for weighted DPO
Built for flexible, scalable RL with HF trainer base!
Check here: github.com/AI4PDLab/Pro...
@aichemist.bsky.social

@aichemist.bsky.social
They however fail to naturally sample rare datapoints, like very high activities.
In our new preprint, we show that RL can solve this without the need for additional data:
arxiv.org/abs/2412.12979
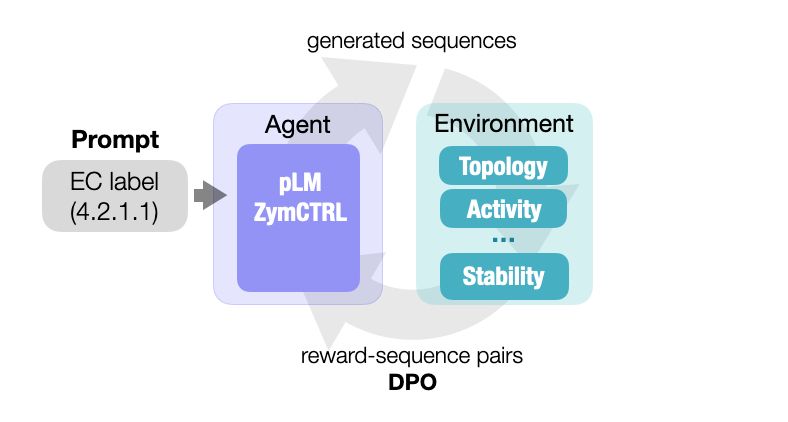
They however fail to naturally sample rare datapoints, like very high activities.
In our new preprint, we show that RL can solve this without the need for additional data:
arxiv.org/abs/2412.12979

