Federico Teloni
@fedeteloni.bsky.social
Postdoc fellow in the @gerlichlab.bsky.social • Genomic integrity and nuclear processes • He/him
https://orcid.org/0000-0003-0953-8010
https://orcid.org/0000-0003-0953-8010
Congratulations for the amazing work!
May 21, 2025 at 9:23 PM
Congratulations for the amazing work!
Fixed link www.biorxiv.org/content/10.1...

Proximity determines donor candidacy during DNA double-stranded break homology directed repair
DNA double-stranded breaks (DSBs) are especially toxic events that can be reversed by homology-directed repair (HDR), wherein information is copied from an intact template molecule. RAD51 mediates ini...
www.biorxiv.org
February 12, 2025 at 9:31 PM
Fixed link www.biorxiv.org/content/10.1...
Yes, after replication sisters are linked by cohesion, making them the preferred template for HR: this restricts the area of search, but additionally we observe that loop-forming and cohesive cohesin work together to define a proper search domain and establish productive contacts
February 12, 2025 at 9:27 PM
Yes, after replication sisters are linked by cohesion, making them the preferred template for HR: this restricts the area of search, but additionally we observe that loop-forming and cohesive cohesin work together to define a proper search domain and establish productive contacts
The study is specific for homologous search of HR during S/G2 phase, we didn't take a look at meiosis yet, but definitely something interesting to pursue.
(Thanks for the DOI heads up, bluesky cut in the preview and it went unnoticed)
(Thanks for the DOI heads up, bluesky cut in the preview and it went unnoticed)
February 12, 2025 at 9:14 PM
The study is specific for homologous search of HR during S/G2 phase, we didn't take a look at meiosis yet, but definitely something interesting to pursue.
(Thanks for the DOI heads up, bluesky cut in the preview and it went unnoticed)
(Thanks for the DOI heads up, bluesky cut in the preview and it went unnoticed)
How does cohesin guide homology search? Preprint from @albertomarin.bsky.social in the Ha lab! https://doi.org/10.1101/2025.02.10.637451
New method for monitoring homology search (RaPID-seq)? Preprint from @charlesyeh.bsky.social in @jcornlab.bsky.social! https://doi.org/10.1101/2025.02.10.63716
New method for monitoring homology search (RaPID-seq)? Preprint from @charlesyeh.bsky.social in @jcornlab.bsky.social! https://doi.org/10.1101/2025.02.10.63716
February 12, 2025 at 5:15 PM
How does cohesin guide homology search? Preprint from @albertomarin.bsky.social in the Ha lab! https://doi.org/10.1101/2025.02.10.637451
New method for monitoring homology search (RaPID-seq)? Preprint from @charlesyeh.bsky.social in @jcornlab.bsky.social! https://doi.org/10.1101/2025.02.10.63716
New method for monitoring homology search (RaPID-seq)? Preprint from @charlesyeh.bsky.social in @jcornlab.bsky.social! https://doi.org/10.1101/2025.02.10.63716
Huge thanks to my co-authors, @imbavienna.bsky.social and the fantastic facilities at @viennabiocenter.bsky.social. This work was supported by @erc.europa.eu, #MSCA, @fwf-at.bsky.social, @snsf-ch.bsky.social, @embo.org, @wwtf.at & #BIF.
For more great science on this topic check the post below 👇
For more great science on this topic check the post below 👇
February 12, 2025 at 5:15 PM
Huge thanks to my co-authors, @imbavienna.bsky.social and the fantastic facilities at @viennabiocenter.bsky.social. This work was supported by @erc.europa.eu, #MSCA, @fwf-at.bsky.social, @snsf-ch.bsky.social, @embo.org, @wwtf.at & #BIF.
For more great science on this topic check the post below 👇
For more great science on this topic check the post below 👇
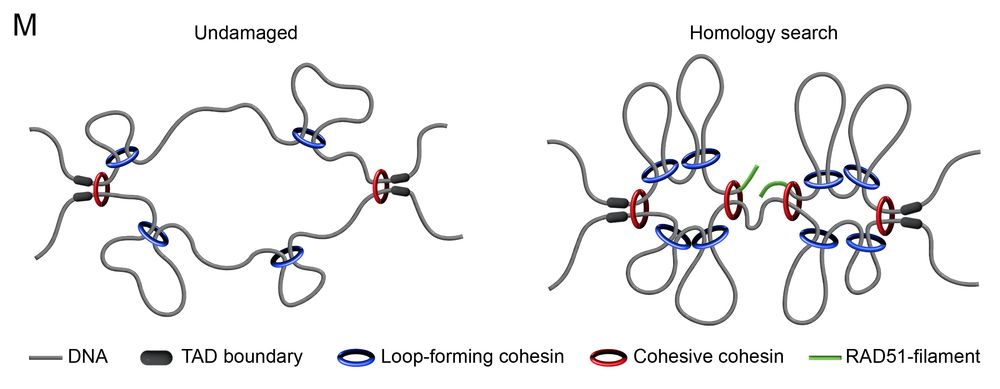
We propose a dual function of cohesin in homology search:
🔹Loop-forming cohesin defines search space, limited by TADs
🔹Cohesive cohesin tethers the break to its sister, to favour productive interactions
This ensures efficient & accurate homology search!
🔹Loop-forming cohesin defines search space, limited by TADs
🔹Cohesive cohesin tethers the break to its sister, to favour productive interactions
This ensures efficient & accurate homology search!
February 12, 2025 at 5:15 PM
We propose a dual function of cohesin in homology search:
🔹Loop-forming cohesin defines search space, limited by TADs
🔹Cohesive cohesin tethers the break to its sister, to favour productive interactions
This ensures efficient & accurate homology search!
🔹Loop-forming cohesin defines search space, limited by TADs
🔹Cohesive cohesin tethers the break to its sister, to favour productive interactions
This ensures efficient & accurate homology search!
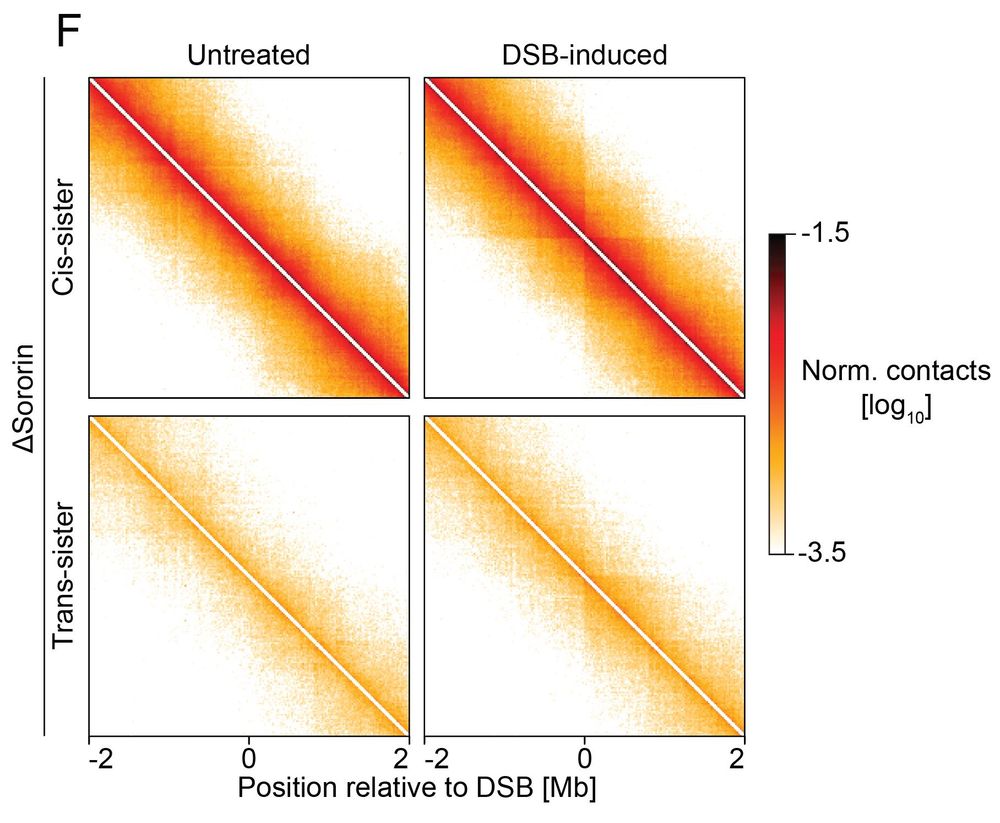
Cohesion directs homology search towards sister chromatid
What happens if we disrupt sister cohesion? Upon sororin depletion (cohesion stabilizer):
✅ Loops remain intact
❌ Contacts between sister chromatids are lost
What happens if we disrupt sister cohesion? Upon sororin depletion (cohesion stabilizer):
✅ Loops remain intact
❌ Contacts between sister chromatids are lost
February 12, 2025 at 5:15 PM
Cohesion directs homology search towards sister chromatid
What happens if we disrupt sister cohesion? Upon sororin depletion (cohesion stabilizer):
✅ Loops remain intact
❌ Contacts between sister chromatids are lost
What happens if we disrupt sister cohesion? Upon sororin depletion (cohesion stabilizer):
✅ Loops remain intact
❌ Contacts between sister chromatids are lost
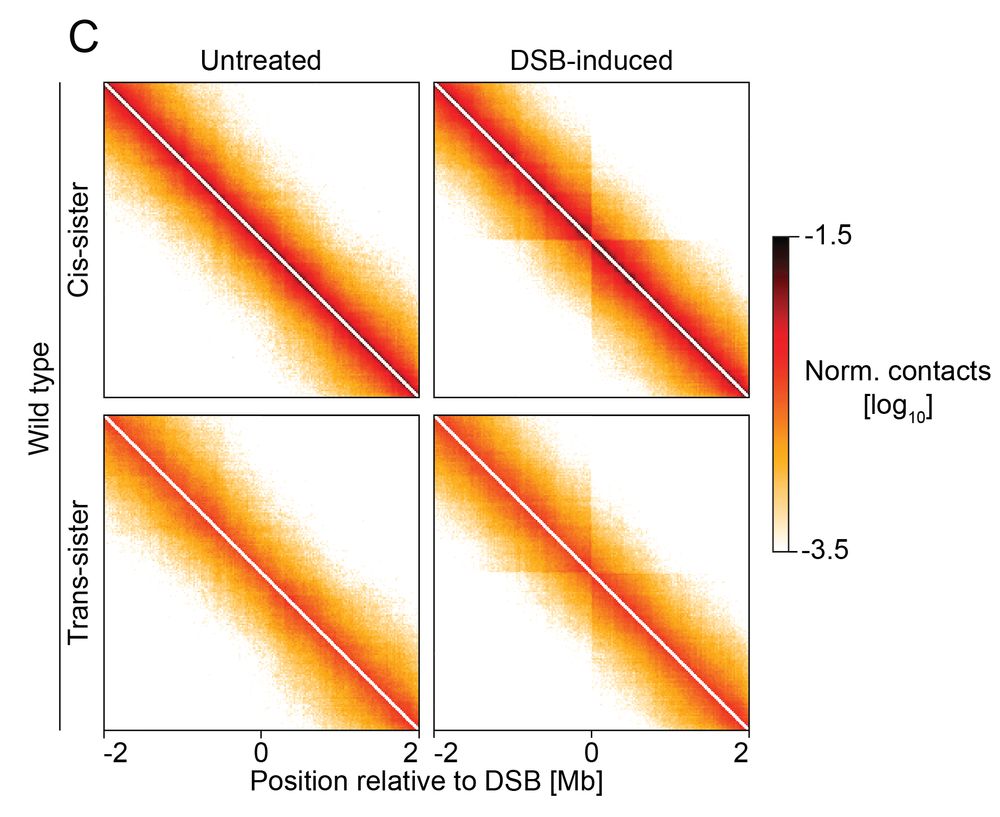
DSBs remodel sister chromatids in cis and trans
Sister-pore-C allows us to track how DNA architecture changes before and after DSBs. We found that DSBs locally increase loops and interactions between the broken ends and the intact sister chromatid.
Sister-pore-C allows us to track how DNA architecture changes before and after DSBs. We found that DSBs locally increase loops and interactions between the broken ends and the intact sister chromatid.
February 12, 2025 at 5:15 PM
DSBs remodel sister chromatids in cis and trans
Sister-pore-C allows us to track how DNA architecture changes before and after DSBs. We found that DSBs locally increase loops and interactions between the broken ends and the intact sister chromatid.
Sister-pore-C allows us to track how DNA architecture changes before and after DSBs. We found that DSBs locally increase loops and interactions between the broken ends and the intact sister chromatid.
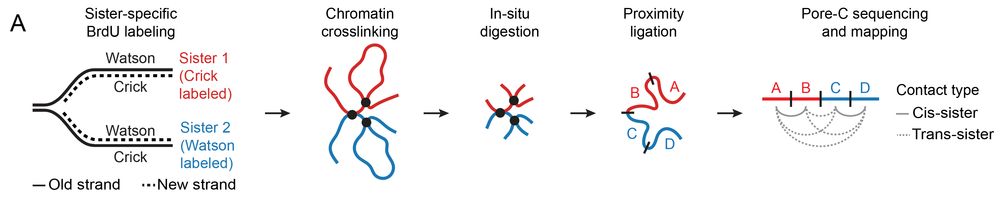
A new tool: sister-pore-C
Homology-directed repair normally uses the sister chromatid as a template, but how does a DSB find its sister locus in 3D space? We developed sister-pore-C, a high-resolution method to map chromatin interactions within & between sister chromatids.
Homology-directed repair normally uses the sister chromatid as a template, but how does a DSB find its sister locus in 3D space? We developed sister-pore-C, a high-resolution method to map chromatin interactions within & between sister chromatids.
February 12, 2025 at 5:15 PM
A new tool: sister-pore-C
Homology-directed repair normally uses the sister chromatid as a template, but how does a DSB find its sister locus in 3D space? We developed sister-pore-C, a high-resolution method to map chromatin interactions within & between sister chromatids.
Homology-directed repair normally uses the sister chromatid as a template, but how does a DSB find its sister locus in 3D space? We developed sister-pore-C, a high-resolution method to map chromatin interactions within & between sister chromatids.
Cohesin-mediated loops control search range
When we disrupted cohesin regulators, search dynamics changed:
🔹 Reducing loops by NIPBL depletion narrowed homology search
🔹 Expanding loops by WAPL depletion made the search broader
Cohesin loops regulate homology search range!
When we disrupted cohesin regulators, search dynamics changed:
🔹 Reducing loops by NIPBL depletion narrowed homology search
🔹 Expanding loops by WAPL depletion made the search broader
Cohesin loops regulate homology search range!

February 12, 2025 at 5:15 PM
Cohesin-mediated loops control search range
When we disrupted cohesin regulators, search dynamics changed:
🔹 Reducing loops by NIPBL depletion narrowed homology search
🔹 Expanding loops by WAPL depletion made the search broader
Cohesin loops regulate homology search range!
When we disrupted cohesin regulators, search dynamics changed:
🔹 Reducing loops by NIPBL depletion narrowed homology search
🔹 Expanding loops by WAPL depletion made the search broader
Cohesin loops regulate homology search range!
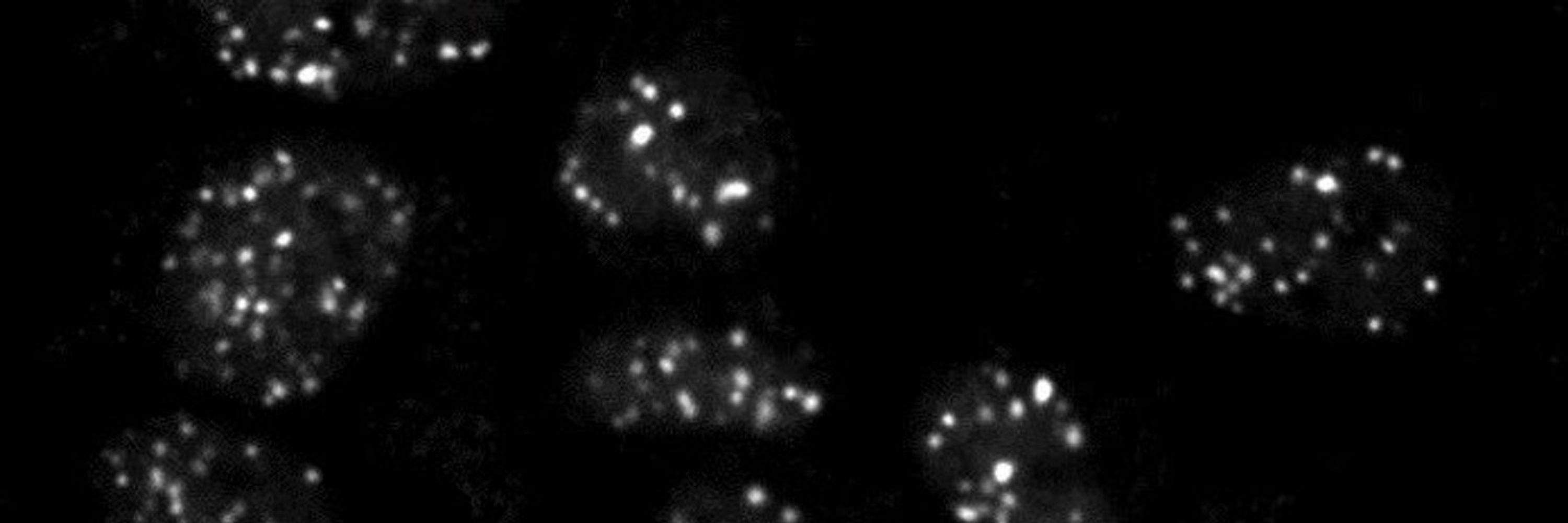
RAD51 filaments sample local TADs
By mapping RAD51, a key repair protein, we found that homology search occurs within topologically associated domains near the breakrather than across the entire genome.
How does chromosome organization influence the search range?
By mapping RAD51, a key repair protein, we found that homology search occurs within topologically associated domains near the breakrather than across the entire genome.
How does chromosome organization influence the search range?

February 12, 2025 at 5:15 PM
RAD51 filaments sample local TADs
By mapping RAD51, a key repair protein, we found that homology search occurs within topologically associated domains near the breakrather than across the entire genome.
How does chromosome organization influence the search range?
By mapping RAD51, a key repair protein, we found that homology search occurs within topologically associated domains near the breakrather than across the entire genome.
How does chromosome organization influence the search range?
Site-specific induction of homology-directed repair
To study how chromosome architecture influences homology search, we induced DSBs at specific genomic sites in S/G2-synchronized human cells. Most breaks were repaired by homologous recombination.
To study how chromosome architecture influences homology search, we induced DSBs at specific genomic sites in S/G2-synchronized human cells. Most breaks were repaired by homologous recombination.

February 12, 2025 at 5:15 PM
Site-specific induction of homology-directed repair
To study how chromosome architecture influences homology search, we induced DSBs at specific genomic sites in S/G2-synchronized human cells. Most breaks were repaired by homologous recombination.
To study how chromosome architecture influences homology search, we induced DSBs at specific genomic sites in S/G2-synchronized human cells. Most breaks were repaired by homologous recombination.
The challenge of homology search
DNA double-strand breaks (DSBs) are dangerous, but homologous recombination repairs them accurately using a homologous template. How do broken DNA ends find homology sites in the folded chromatin architecture of the nucleus?
DNA double-strand breaks (DSBs) are dangerous, but homologous recombination repairs them accurately using a homologous template. How do broken DNA ends find homology sites in the folded chromatin architecture of the nucleus?
February 12, 2025 at 5:15 PM
The challenge of homology search
DNA double-strand breaks (DSBs) are dangerous, but homologous recombination repairs them accurately using a homologous template. How do broken DNA ends find homology sites in the folded chromatin architecture of the nucleus?
DNA double-strand breaks (DSBs) are dangerous, but homologous recombination repairs them accurately using a homologous template. How do broken DNA ends find homology sites in the folded chromatin architecture of the nucleus?
You never know what people might find valuable these days, I guess willhaben is the way!
January 23, 2025 at 10:17 AM
You never know what people might find valuable these days, I guess willhaben is the way!

