https://orcid.org/0000-0003-0953-8010
🔹Loop-forming cohesin defines search space, limited by TADs
🔹Cohesive cohesin tethers the break to its sister, to favour productive interactions
This ensures efficient & accurate homology search!
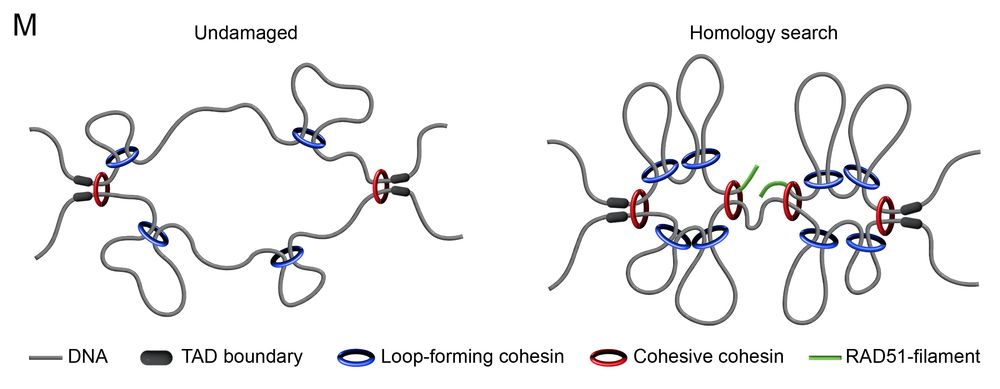
🔹Loop-forming cohesin defines search space, limited by TADs
🔹Cohesive cohesin tethers the break to its sister, to favour productive interactions
This ensures efficient & accurate homology search!
What happens if we disrupt sister cohesion? Upon sororin depletion (cohesion stabilizer):
✅ Loops remain intact
❌ Contacts between sister chromatids are lost
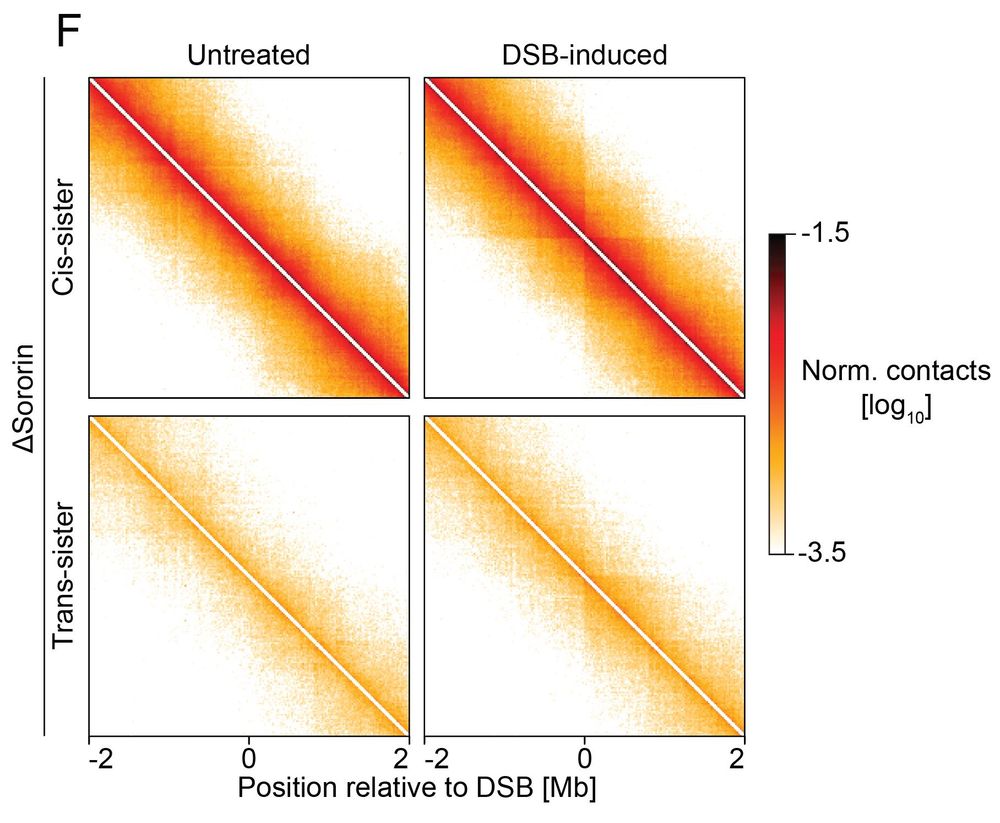
What happens if we disrupt sister cohesion? Upon sororin depletion (cohesion stabilizer):
✅ Loops remain intact
❌ Contacts between sister chromatids are lost
Sister-pore-C allows us to track how DNA architecture changes before and after DSBs. We found that DSBs locally increase loops and interactions between the broken ends and the intact sister chromatid.
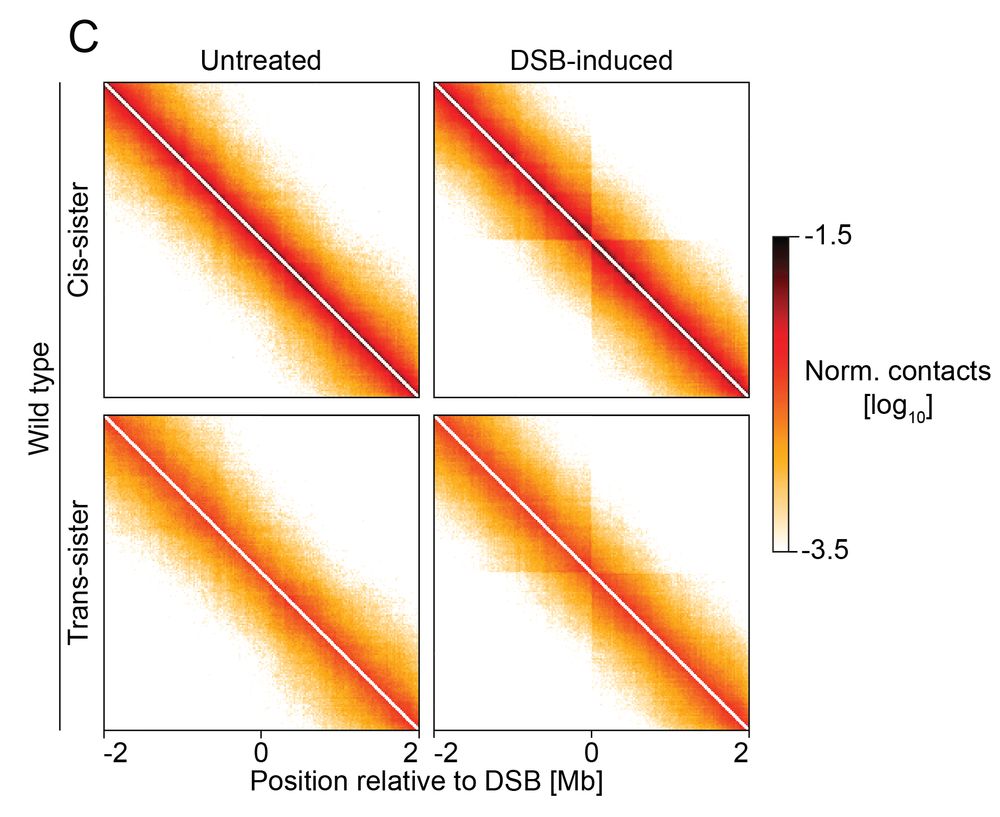
Sister-pore-C allows us to track how DNA architecture changes before and after DSBs. We found that DSBs locally increase loops and interactions between the broken ends and the intact sister chromatid.
Homology-directed repair normally uses the sister chromatid as a template, but how does a DSB find its sister locus in 3D space? We developed sister-pore-C, a high-resolution method to map chromatin interactions within & between sister chromatids.
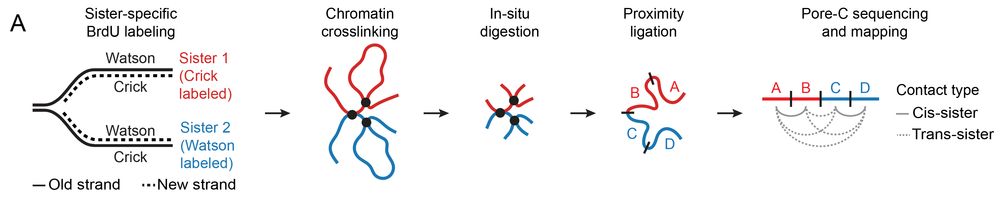
Homology-directed repair normally uses the sister chromatid as a template, but how does a DSB find its sister locus in 3D space? We developed sister-pore-C, a high-resolution method to map chromatin interactions within & between sister chromatids.
When we disrupted cohesin regulators, search dynamics changed:
🔹 Reducing loops by NIPBL depletion narrowed homology search
🔹 Expanding loops by WAPL depletion made the search broader
Cohesin loops regulate homology search range!

When we disrupted cohesin regulators, search dynamics changed:
🔹 Reducing loops by NIPBL depletion narrowed homology search
🔹 Expanding loops by WAPL depletion made the search broader
Cohesin loops regulate homology search range!
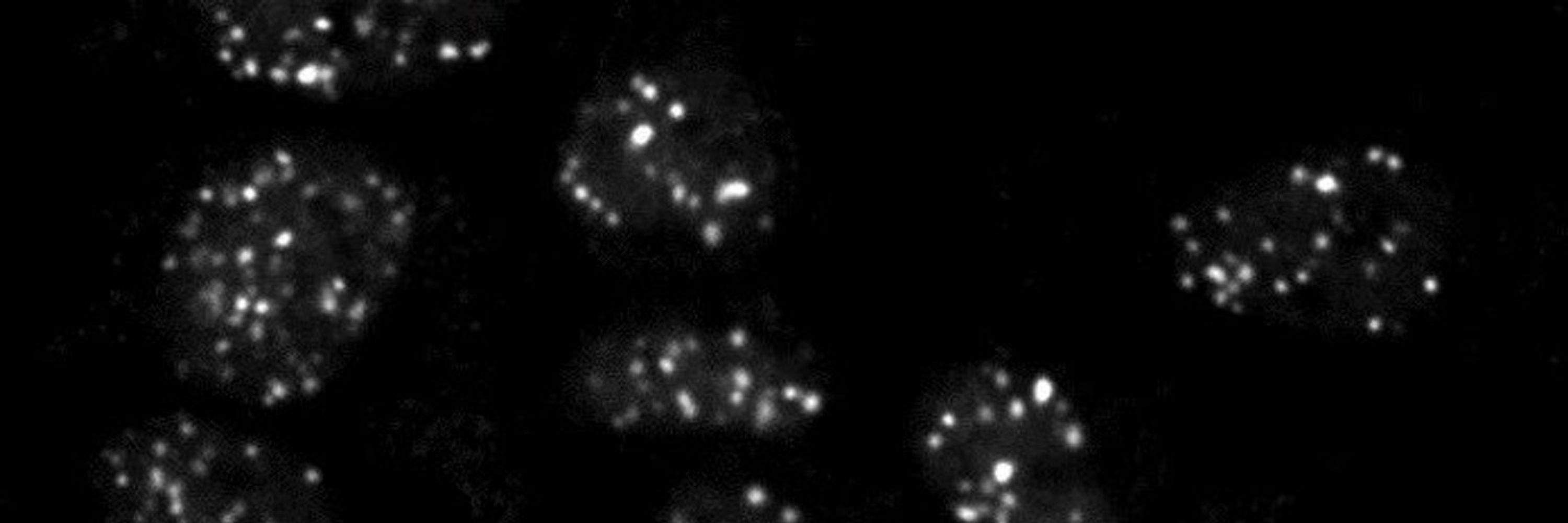
By mapping RAD51, a key repair protein, we found that homology search occurs within topologically associated domains near the breakrather than across the entire genome.
How does chromosome organization influence the search range?

By mapping RAD51, a key repair protein, we found that homology search occurs within topologically associated domains near the breakrather than across the entire genome.
How does chromosome organization influence the search range?
To study how chromosome architecture influences homology search, we induced DSBs at specific genomic sites in S/G2-synchronized human cells. Most breaks were repaired by homologous recombination.

To study how chromosome architecture influences homology search, we induced DSBs at specific genomic sites in S/G2-synchronized human cells. Most breaks were repaired by homologous recombination.

