
---
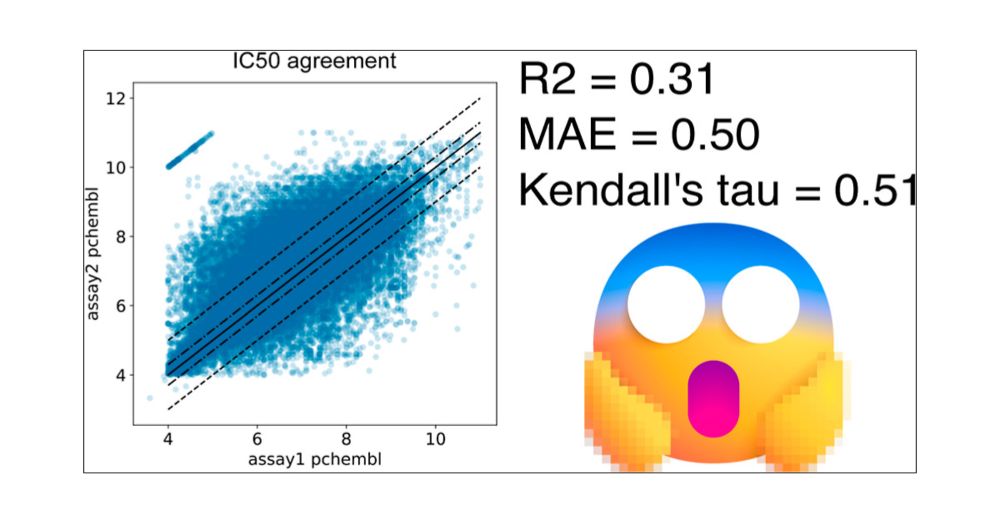
#proteomics #prot-paper
---
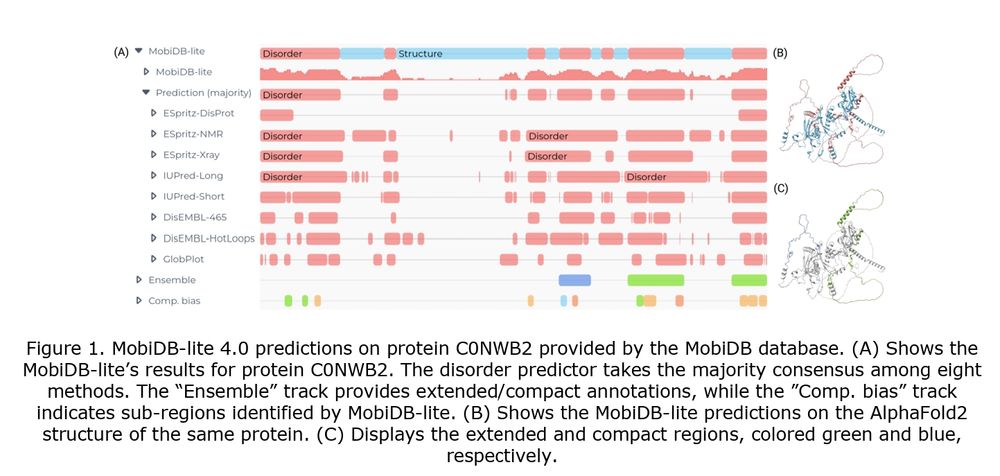
#proteomics #prot-paper
---
#proteomics #prot-preprint

---
#proteomics #prot-preprint
arxiv.org/abs/2410.18403
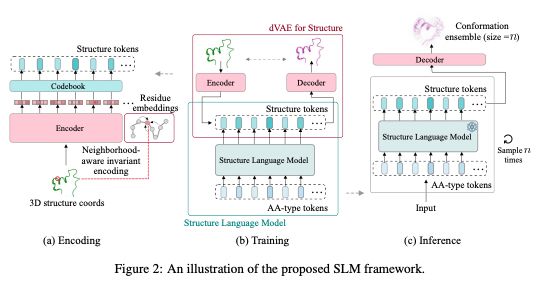
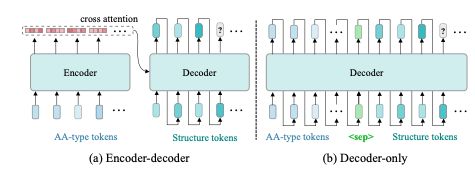
arxiv.org/abs/2410.18403
🌐https://bfvd.foldseek.com
💾https://bfvd.steineggerlab.workers.dev/
1/3
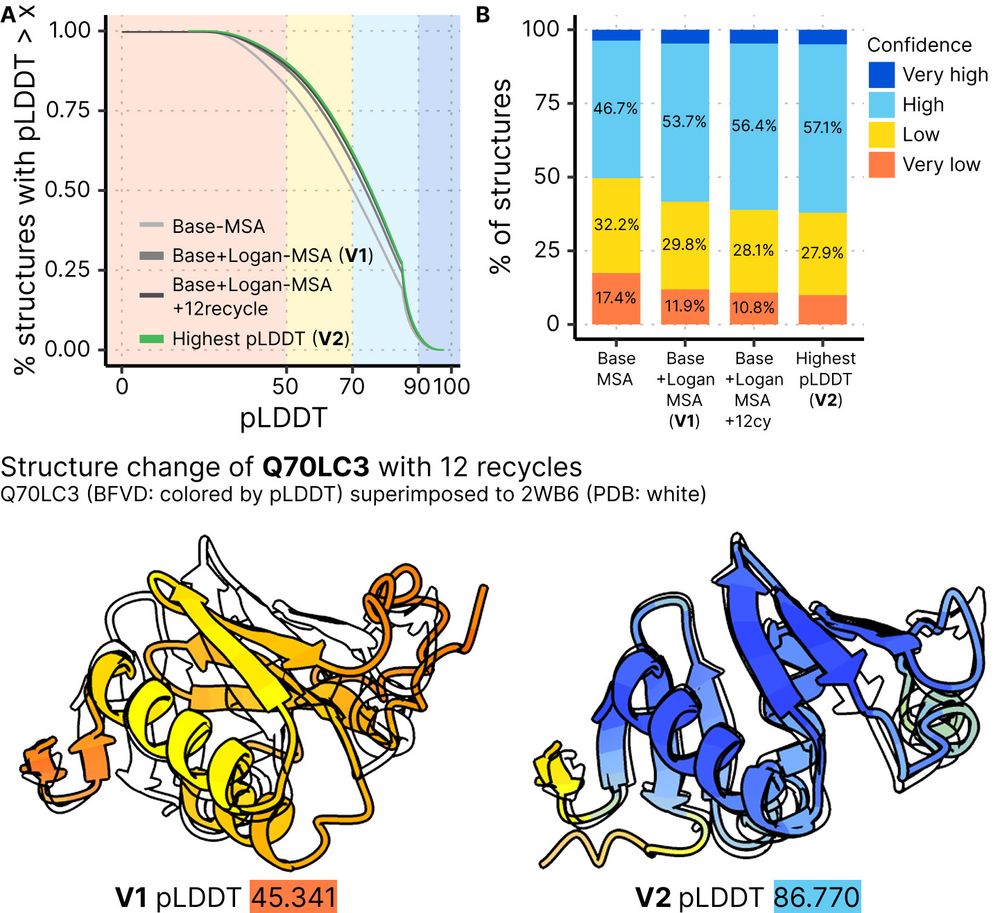
🌐https://bfvd.foldseek.com
💾https://bfvd.steineggerlab.workers.dev/
1/3


