
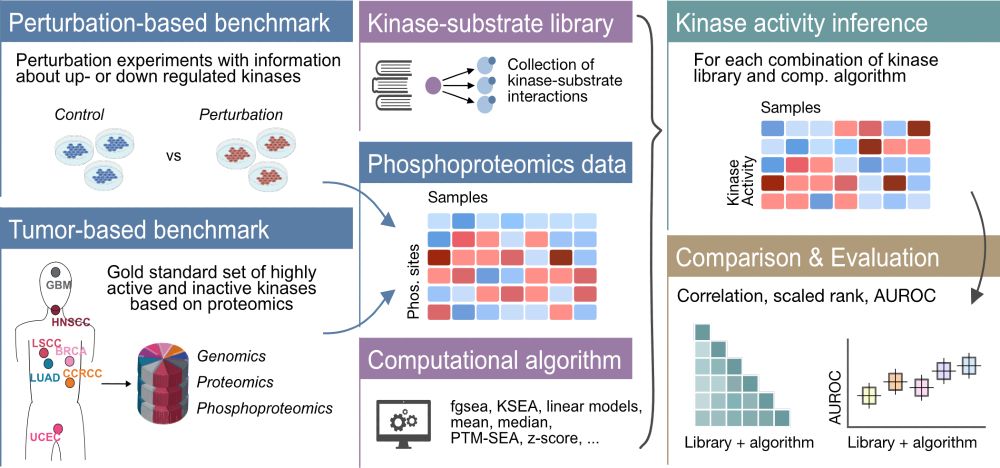
🔗 biorxiv.org/content/10.1...
A short thread (by Juliane Weller)👇

🔗 biorxiv.org/content/10.1...
A short thread (by Juliane Weller)👇
Learn more: www.cell.com/cell/fulltex...

Learn more: www.cell.com/cell/fulltex...
Details & apply by 13/10/25: tinyurl.com/4shdw8dk

Details & apply by 13/10/25: tinyurl.com/4shdw8dk
🔗 Manuscript: www.biorxiv.org/content/10.1...
💻 Code: partipy.readthedocs.io

🔗 Manuscript: www.biorxiv.org/content/10.1...
💻 Code: partipy.readthedocs.io
We will decode and recode tissue ecosystems using AI + big data…
Our amazing faculty include Roser Vento-Tormo, @bayraktarlab.bsky.social, Mo Lotfollahi, Sam Behjati and Song Chen..
sangerinstitute.blog/2025/05/20/i...

We will decode and recode tissue ecosystems using AI + big data…
Our amazing faculty include Roser Vento-Tormo, @bayraktarlab.bsky.social, Mo Lotfollahi, Sam Behjati and Song Chen..
We present our new alignment framework TOAST www.biorxiv.org/content/10.1...
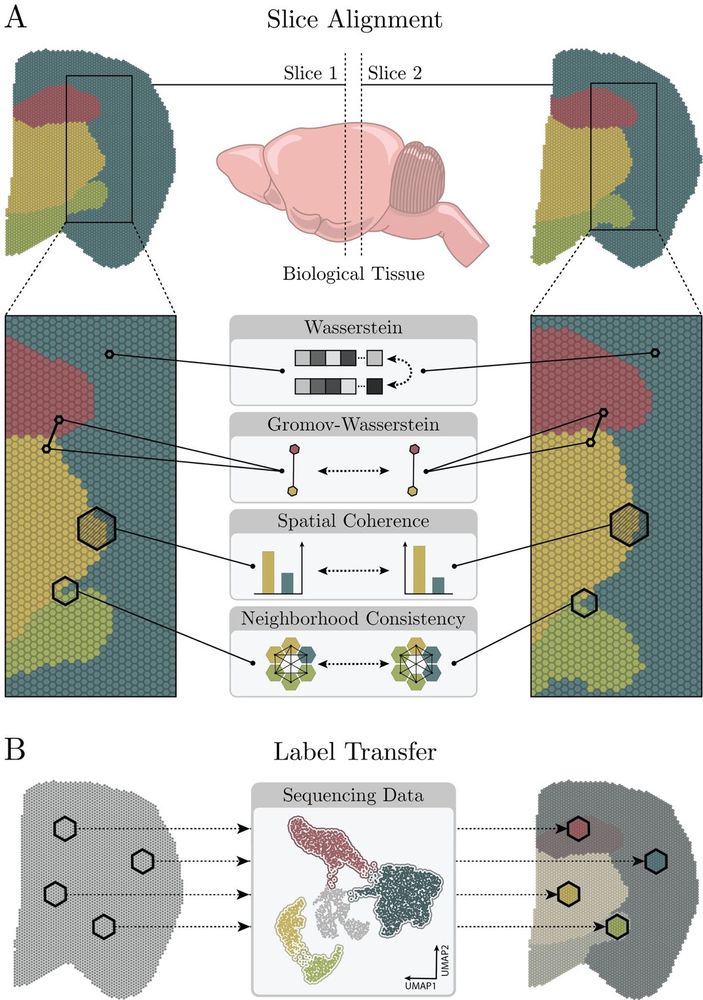
We present our new alignment framework TOAST www.biorxiv.org/content/10.1...
Ever wondered how to best quantify cell-cell neighbor preferences in tissues?
We compared 9+ neighbor preference (NEP) methods for analysing spatial omics data and propose a novel approach that combines the most relevant analysis features which we call COZI 🔬✨
Read more: doi.org/10.1101/2025...
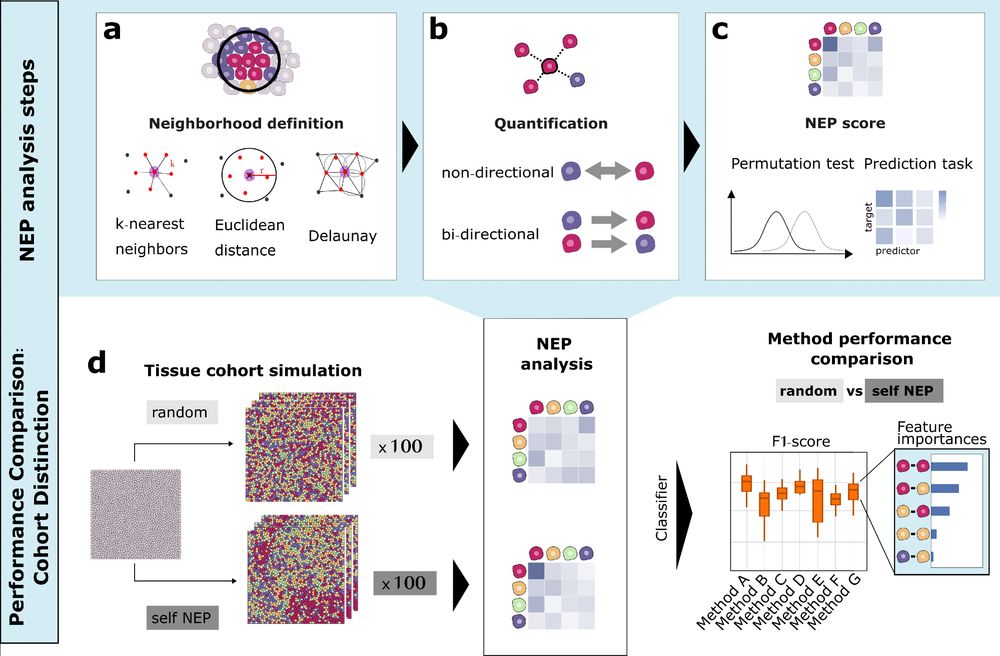
Ever wondered how to best quantify cell-cell neighbor preferences in tissues?
We compared 9+ neighbor preference (NEP) methods for analysing spatial omics data and propose a novel approach that combines the most relevant analysis features which we call COZI 🔬✨
Read more: doi.org/10.1101/2025...
Paper: doi.org/10.1101/2024.12.20.629764
Code: github.com/saezlab/greta
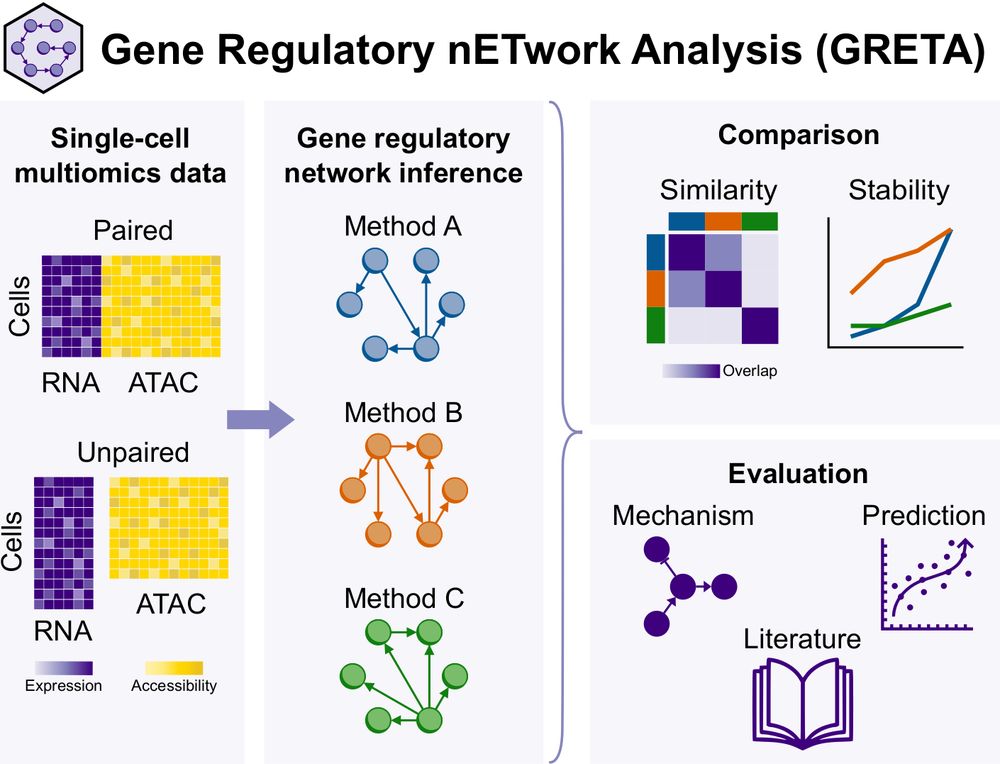
Paper: doi.org/10.1101/2024.12.20.629764
Code: github.com/saezlab/greta
www.biorxiv.org/content/10.1...
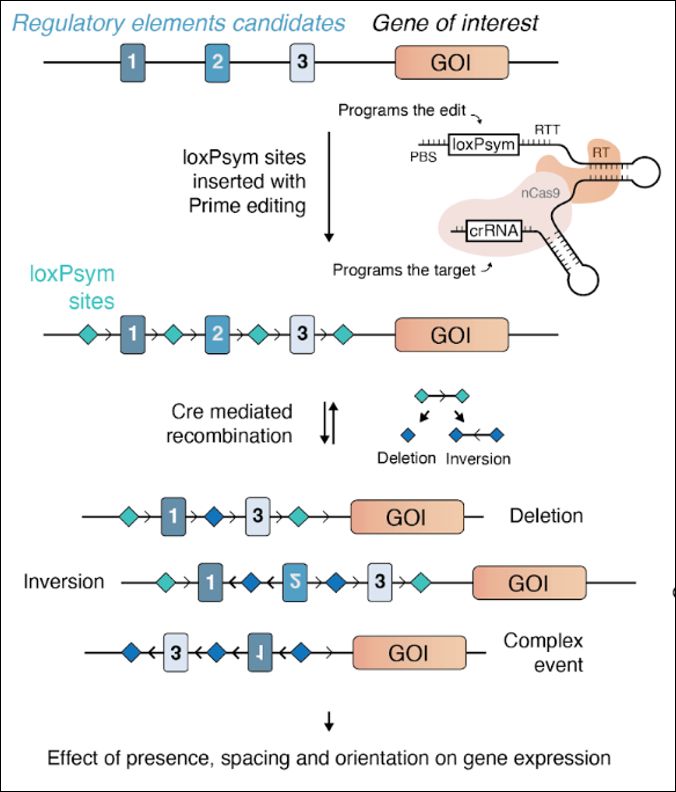
www.biorxiv.org/content/10.1...
Today we published a study on the role of skin B cells in cutaneous T cell lymphoma www.nature.com/articles/s41...
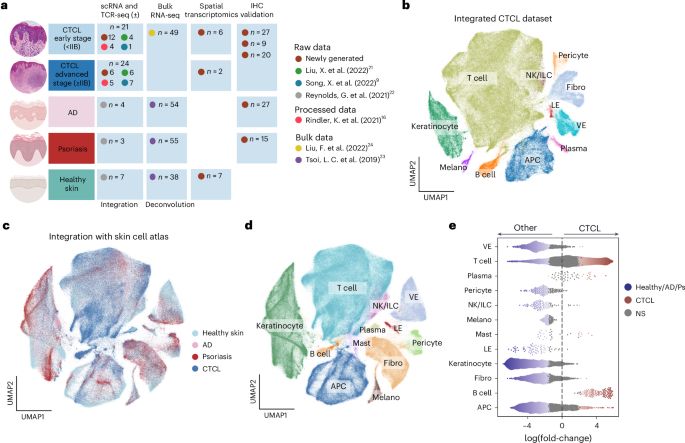
Today we published a study on the role of skin B cells in cutaneous T cell lymphoma www.nature.com/articles/s41...


