Elizabeth Atkinson
@egatkinson.bsky.social
Population and statistical genomicist working to make genomics fully representative. Views are my own. (she/her)
Pinned
📃 We’re excited to share our latest work, now published in Nature Communications — a major update to the Genome Aggregation Database (gnomAD) that improves allele frequency resolution for two gnomAD-defined genetic ancestry groups using local ancestry inference (LAI).
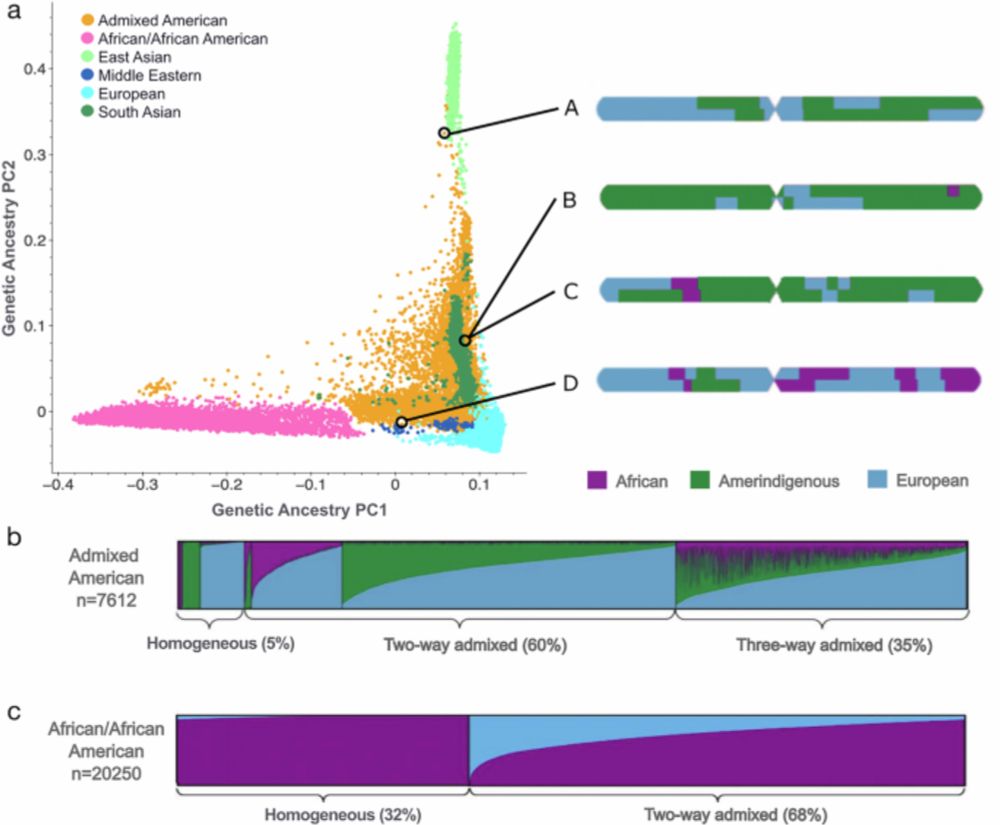
Improved allele frequencies in gnomAD through local ancestry inference - Nature Communications
This study incorporates local ancestry into the Genome Aggregation Database (gnomAD) to improve allele frequency estimates for admixed populations, enhancing variant interpretation and enabling more accurate and equitable genomic research and clinical care.
www.nature.com
Thrilled to share our new @natcomms.nature.com paper on local ancestry informed allele frequencies in gnomAD, which are live now on the browser! Check out my stellar PhD student @pragskore.bsky.social’s Bluetorial on how this brings finer detail to variant interpretation 🧬🖥️
A fun lab outing to the zoo ahead of conference season! 🦒

October 12, 2025 at 2:27 AM
A fun lab outing to the zoo ahead of conference season! 🦒
Reposted by Elizabeth Atkinson
Texas Children's/Baylor College of Medicine Researchers Create Groundbreaking Tool to Improve Accuracy of #GeneticTesting @egatkinson.bsky.social @bcmgenetics.bsky.social @bcmhouston.bsky.social #TCHResearchNews #TexasChildrens @natcomms.nature.com tinyurl.com/jj6kyrrv

October 6, 2025 at 2:16 PM
Texas Children's/Baylor College of Medicine Researchers Create Groundbreaking Tool to Improve Accuracy of #GeneticTesting @egatkinson.bsky.social @bcmgenetics.bsky.social @bcmhouston.bsky.social #TCHResearchNews #TexasChildrens @natcomms.nature.com tinyurl.com/jj6kyrrv
Thrilled to share our new @natcomms.nature.com paper on local ancestry informed allele frequencies in gnomAD, which are live now on the browser! Check out my stellar PhD student @pragskore.bsky.social’s Bluetorial on how this brings finer detail to variant interpretation 🧬🖥️
📃 We’re excited to share our latest work, now published in Nature Communications — a major update to the Genome Aggregation Database (gnomAD) that improves allele frequency resolution for two gnomAD-defined genetic ancestry groups using local ancestry inference (LAI).

Improved allele frequencies in gnomAD through local ancestry inference - Nature Communications
This study incorporates local ancestry into the Genome Aggregation Database (gnomAD) to improve allele frequency estimates for admixed populations, enhancing variant interpretation and enabling more accurate and equitable genomic research and clinical care.
www.nature.com
October 6, 2025 at 6:44 PM
Thrilled to share our new @natcomms.nature.com paper on local ancestry informed allele frequencies in gnomAD, which are live now on the browser! Check out my stellar PhD student @pragskore.bsky.social’s Bluetorial on how this brings finer detail to variant interpretation 🧬🖥️
Reposted by Elizabeth Atkinson
A project many years in the process, we’re pleased to present our work on multi-ancestry meta-analysis across a boatload of traits in the UK Biobank: www.nature.com/articles/s41...

Pan-UK Biobank genome-wide association analyses enhance discovery and resolution of ancestry-enriched effects - Nature Genetics
Genome-wide analyses for 7,266 traits leveraging data from several genetic ancestry groups in UK Biobank identify new associations and enhance resources for interpreting risk variants across diverse p...
www.nature.com
September 18, 2025 at 5:25 PM
A project many years in the process, we’re pleased to present our work on multi-ancestry meta-analysis across a boatload of traits in the UK Biobank: www.nature.com/articles/s41...
Delighted to amplify my talented PhD student’s work! Check it out for a great way to streamline and harmonize Tractor analyses.
🚨 New preprint! Traditional GWAS often exclude admixed individuals, missing ancestry-enriched genetic signals. We built Tractor Nextflow Workflow - a modular, scalable pipeline that automates phasing, LAI, and Tractor GWAS, alongside comprehensive documentation.
Preprint: doi.org/10.1101/2025...
Preprint: doi.org/10.1101/2025...

Tractor Workflow Pipeline: A Scalable Nextflow Framework for Local Ancestry-Aware Genome-Wide Association Studies
The routine exclusion of admixed individuals from traditional Genome-Wide Association Studies (GWAS) due to concerns about spurious associations has hindered genetic analyses involving multiple ancest...
www.biorxiv.org
September 13, 2025 at 12:47 AM
Delighted to amplify my talented PhD student’s work! Check it out for a great way to streamline and harmonize Tractor analyses.
🚨 New perspective piece in @ajhgnews.bsky.social! 🚨
We developed a hands-on training resource for large-scale genomic data analysis in the All of Us Researcher Workbench, now published here:
We developed a hands-on training resource for large-scale genomic data analysis in the All of Us Researcher Workbench, now published here:
📣Online now!
📄Implementing a training resource for large-scale genomic data analysis in the All of Us Researcher Workbench
🧑💻 @jasminebakerphd.bsky.social @egatkinson.bsky.social & co
📄Implementing a training resource for large-scale genomic data analysis in the All of Us Researcher Workbench
🧑💻 @jasminebakerphd.bsky.social @egatkinson.bsky.social & co

Implementing a training resource for large-scale genomic data analysis in the All of Us Researcher Workbench
Baker et al. present a cloud-based training program using the All of Us Researcher
Workbench to teach large-scale genomic data analysis. Early-career researchers gained
hands-on experience in GWASs, q...
www.cell.com
July 22, 2025 at 4:36 PM
🚨 New perspective piece in @ajhgnews.bsky.social! 🚨
We developed a hands-on training resource for large-scale genomic data analysis in the All of Us Researcher Workbench, now published here:
We developed a hands-on training resource for large-scale genomic data analysis in the All of Us Researcher Workbench, now published here:
We're excited to introduce Tractor-Mix, our new method for GWAS in admixed cohorts with relatedness, led by the fantastic @doubletaotan.bsky.social! Read the full preprint here: www.medrxiv.org/content/10.1...
Thanks to all our amazing collaborators who helped make this work possible!
Thanks to all our amazing collaborators who helped make this work possible!

June 9, 2025 at 6:31 PM
We're excited to introduce Tractor-Mix, our new method for GWAS in admixed cohorts with relatedness, led by the fantastic @doubletaotan.bsky.social! Read the full preprint here: www.medrxiv.org/content/10.1...
Thanks to all our amazing collaborators who helped make this work possible!
Thanks to all our amazing collaborators who helped make this work possible!
Check out my stellar PhD student, Pragati's talk on our work generating local ancestry informed frequency estimates in gnomAD as part of the prestigious Emerging Genomic Scientist Symposium next week! Congrats on being selected for this amazing event!
Human Genetics | Genomic Scientist Fellows | UCLA Medical School
The Emerging Genomic Scientist Fellows Program is a cornerstone of justice, equity, diversity, and inclusion initiatives in the Department of Human Genetics.
medschool.ucla.edu
April 9, 2025 at 3:57 PM
Check out my stellar PhD student, Pragati's talk on our work generating local ancestry informed frequency estimates in gnomAD as part of the prestigious Emerging Genomic Scientist Symposium next week! Congrats on being selected for this amazing event!
I'm delighted to be part of this symposium, put on by University of Pennsylvania Perelman School of Medicine, and led by @bpasaniuc.bsky.social and @sarahtishkoff.bsky.social. See you in a few weeks! upenn.co1.qualtrics.com/jfe/form/SV_...

April 3, 2025 at 2:48 PM
I'm delighted to be part of this symposium, put on by University of Pennsylvania Perelman School of Medicine, and led by @bpasaniuc.bsky.social and @sarahtishkoff.bsky.social. See you in a few weeks! upenn.co1.qualtrics.com/jfe/form/SV_...
📃 I am excited to share our @naturegenet.bsky.social review paper on the state of psychiatric genetics in Latin America, put forth by the Latin American Genomics Consortium (LAGC)! 🎉
www.nature.com/articles/s41...
www.nature.com/articles/s41...

Psychiatric genetics in the diverse landscape of Latin American populations - Nature Genetics
Latin America and the Caribbean remain largely underrepresented in psychiatric genetics research. This Review highlights the need for more research in these populations to advance genetic insights and...
www.nature.com
April 2, 2025 at 3:24 PM
📃 I am excited to share our @naturegenet.bsky.social review paper on the state of psychiatric genetics in Latin America, put forth by the Latin American Genomics Consortium (LAGC)! 🎉
www.nature.com/articles/s41...
www.nature.com/articles/s41...
www.biorxiv.org/content/10.1...
📄 New preprint alert! 📄
@helenyslin.bsky.social benchmarked polygenic scores (PGS) using genotyping arrays vs whole-genome sequencing (WGS) across traits & populations in @AllofUsResearch. What’s the best approach? The answer is… it depends! 🧵⬇️
📄 New preprint alert! 📄
@helenyslin.bsky.social benchmarked polygenic scores (PGS) using genotyping arrays vs whole-genome sequencing (WGS) across traits & populations in @AllofUsResearch. What’s the best approach? The answer is… it depends! 🧵⬇️

Differential performance of polygenic prediction across traits and populations depending on genotype discovery approach
Polygenic scores (PGS) are widely used for estimating genetic predisposition to complex traits by aggregating the effects of common variants into a single measure. They hold promise in identifying ind...
www.biorxiv.org
March 20, 2025 at 2:53 PM
www.biorxiv.org/content/10.1...
📄 New preprint alert! 📄
@helenyslin.bsky.social benchmarked polygenic scores (PGS) using genotyping arrays vs whole-genome sequencing (WGS) across traits & populations in @AllofUsResearch. What’s the best approach? The answer is… it depends! 🧵⬇️
📄 New preprint alert! 📄
@helenyslin.bsky.social benchmarked polygenic scores (PGS) using genotyping arrays vs whole-genome sequencing (WGS) across traits & populations in @AllofUsResearch. What’s the best approach? The answer is… it depends! 🧵⬇️
A fun Atkinson lab meeting with 100% membership in person! Exciting to see the whole group together

March 20, 2025 at 2:30 PM
A fun Atkinson lab meeting with 100% membership in person! Exciting to see the whole group together
Reposted by Elizabeth Atkinson
TODAY! @standupforscience.bsky.social is taking place in DC & across the U.S. to help protect the American scientific enterprise & defend science as a public good that drives social, political, & economic progress. Find your local site: buff.ly/3EWhSZM
#scienceforall #advocacy 1/2🧵
#scienceforall #advocacy 1/2🧵

March 7, 2025 at 2:09 PM
TODAY! @standupforscience.bsky.social is taking place in DC & across the U.S. to help protect the American scientific enterprise & defend science as a public good that drives social, political, & economic progress. Find your local site: buff.ly/3EWhSZM
#scienceforall #advocacy 1/2🧵
#scienceforall #advocacy 1/2🧵
So proud of my amazing lab member @jhmauer.bsky.social being featured by AJHG for this fantastic work!
In Feb, The American Journal of Human Genetics sat with Jessica Honoratu-Mauer to discuss her recent paper, “Characterizing features affecting local ancestry inference performance in admixed populations.” To read the interview visit www.ashg.org/ajhg/inside-... Congrats Jessica on your publication!

Inside AJHG: A Chat with Jessica Honorato-Mauer
Editors of The American Journal of Human Genetics sat down with Jessica Honorato-Mauer, MS to discuss their recently published paper “Characterizing features affecting local ancestry inference perform...
www.ashg.org
March 6, 2025 at 6:54 PM
So proud of my amazing lab member @jhmauer.bsky.social being featured by AJHG for this fantastic work!
Reposted by Elizabeth Atkinson
We asked three chairs of PGC Working Groups: what is the goal for their research this year? @niamhmullins.bsky.social, Barbara Franke and @egatkinson.bsky.social #womeninSTEM #InternationalWomensDay2025



March 6, 2025 at 6:40 PM
We asked three chairs of PGC Working Groups: what is the goal for their research this year? @niamhmullins.bsky.social, Barbara Franke and @egatkinson.bsky.social #womeninSTEM #InternationalWomensDay2025
Reposted by Elizabeth Atkinson
Grace Tietz is a Ph.D. student in the Genetics & Genomics Program at Baylor in the lab of Dr. Elizabeth Atkinson's. Grace's primary project aims to improve genetic testing for cardiovascular disease across populations. She was just awarded her F31 from the NHLBI.

February 12, 2025 at 12:48 AM
Grace Tietz is a Ph.D. student in the Genetics & Genomics Program at Baylor in the lab of Dr. Elizabeth Atkinson's. Grace's primary project aims to improve genetic testing for cardiovascular disease across populations. She was just awarded her F31 from the NHLBI.

