
Researcher at UP Olomouc.
www.nature.com/articles/d41...

www.nature.com/articles/d41...







"k-mer approaches for biodiversity genomics"
We discuss k-mer spectra, sex determination, allopolyploid subgenome separation, among other topics - and offer a large tutorial list
Link to paper:
genome.cshlp.org/content/earl...
Link to all tutorials:
github.com/KamilSJaron/...



"k-mer approaches for biodiversity genomics"
We discuss k-mer spectra, sex determination, allopolyploid subgenome separation, among other topics - and offer a large tutorial list
Link to paper:
genome.cshlp.org/content/earl...
Link to all tutorials:
github.com/KamilSJaron/...

🔗 doi.org/10.1093/jeb/voae136
🔗 doi.org/10.1093/jeb/voae136

Earlier that day:

Earlier that day:
pnas.org/doi/10.1073/...

pnas.org/doi/10.1073/...
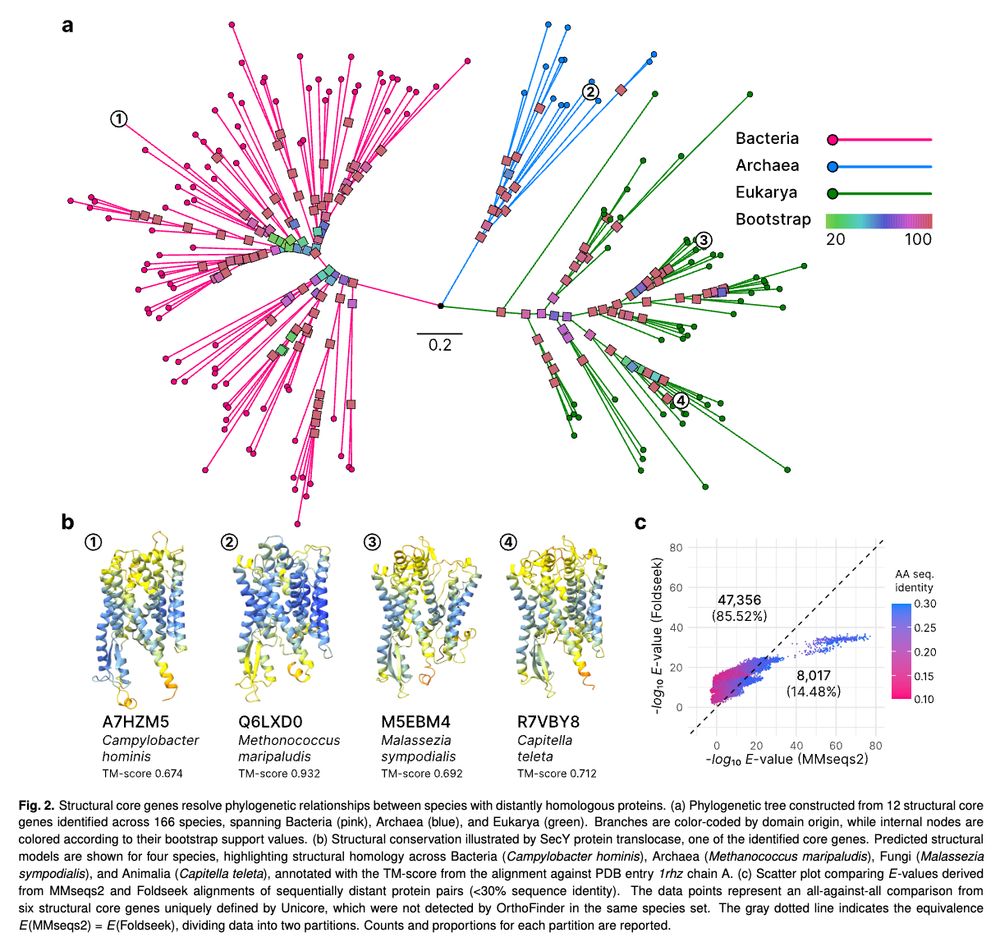
If so, the one thing you all can do is persuade colleagues to submit and make this a norm. 1/2
If so, the one thing you all can do is persuade colleagues to submit and make this a norm. 1/2
@lucalivraghi.bsky.social
www.biorxiv.org/content/10.1...
We mapped optix (again?!) as the switch gene of a natural polymorphism, this time controlling silver patches of a mountain butterfly. Gorgeous RNAi validation, evidence of selective sweeps, introgression
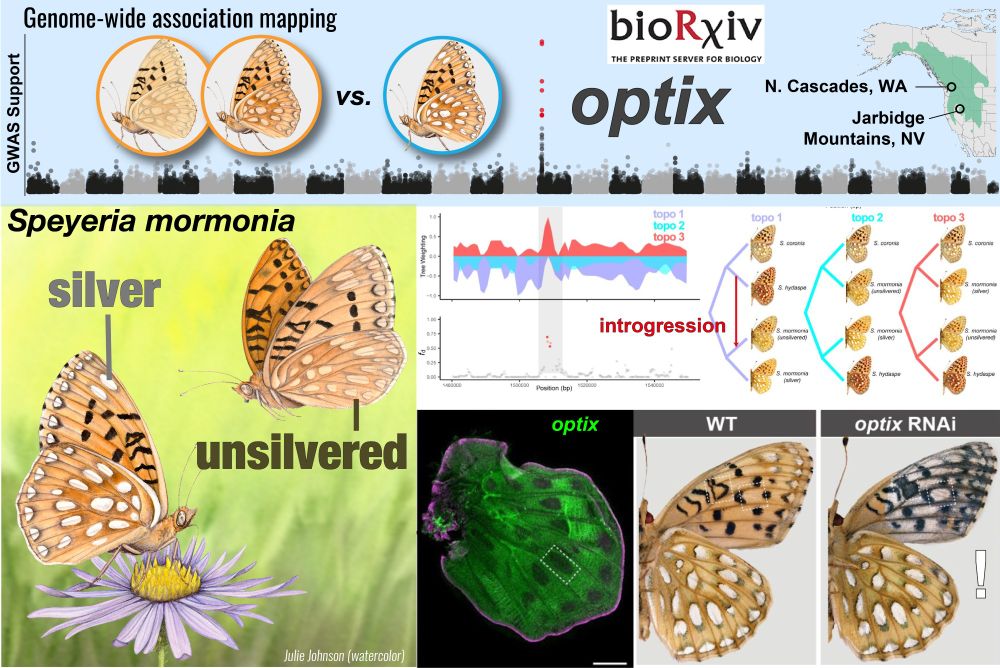
@lucalivraghi.bsky.social
www.biorxiv.org/content/10.1...
We mapped optix (again?!) as the switch gene of a natural polymorphism, this time controlling silver patches of a mountain butterfly. Gorgeous RNAi validation, evidence of selective sweeps, introgression
You can use paralogs to reduce long-branch attraction!
This will be especially helpful when there are no extant taxa that can be sampled to break up these branches.
www.biorxiv.org/content/10.1...

You can use paralogs to reduce long-branch attraction!
This will be especially helpful when there are no extant taxa that can be sampled to break up these branches.
www.biorxiv.org/content/10.1...

peerj.com/articles/185...
⚒️🧪🦀🦑 #evosky

peerj.com/articles/185...
⚒️🧪🦀🦑 #evosky



