

Engineering E. coli bacteria to turn plastic waste into paracetamol (Tylenol)
www.nature.com/articles/s41...
www.nature.com/articles/s41...
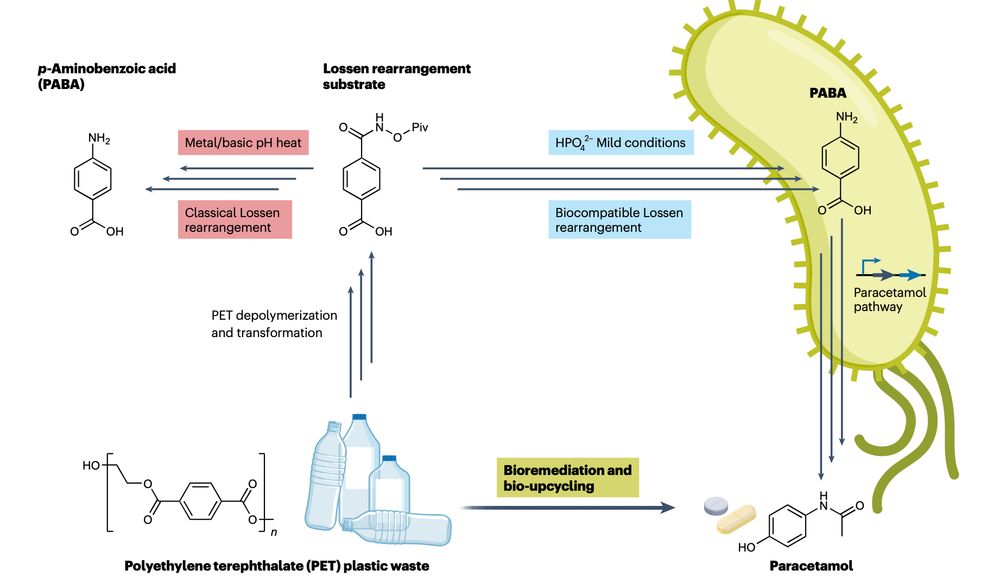
Engineering E. coli bacteria to turn plastic waste into paracetamol (Tylenol)
www.nature.com/articles/s41...
www.nature.com/articles/s41...
g-xTB marks not just an evolution, but a revolution in the capabilities of semiempirical quantum chemistry. Convince yourself! A thread.
🔗 chemrxiv.org/engage/chemr...
#compchem

g-xTB marks not just an evolution, but a revolution in the capabilities of semiempirical quantum chemistry. Convince yourself! A thread.
🔗 chemrxiv.org/engage/chemr...
#compchem

