We used genomic barcoding + scRNAseq in chick & human embryos to reveal a lineage architecture that reshapes how we understand neural tube development & cell fate decisions
🧵👇
www.biorxiv.org/content/10.1...

We used genomic barcoding + scRNAseq in chick & human embryos to reveal a lineage architecture that reshapes how we understand neural tube development & cell fate decisions
🧵👇
www.biorxiv.org/content/10.1...
Absolutely loved joint supervision w. Zena! Perfect @crick.ac.uk & @kingsioppn.bsky.social combo
Dana’s day had a stiff competition 😉😉😉
Can't wait for what's next!


Can't wait for what's next!
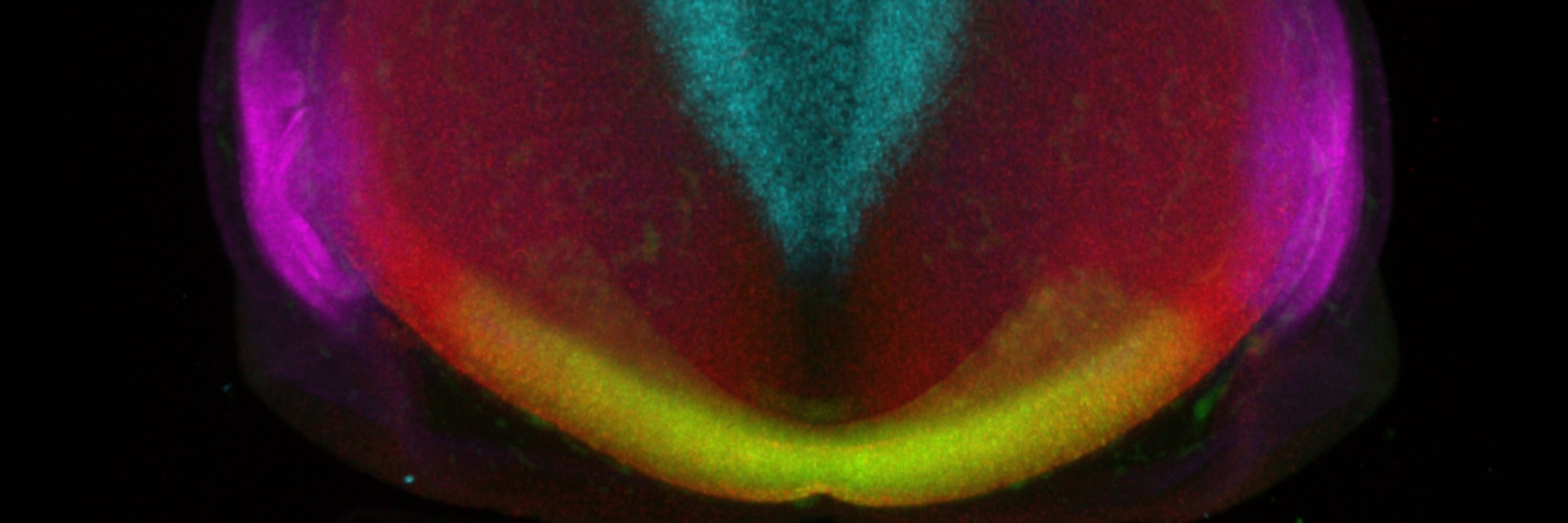
This week, it's a 3D image of a multiplex, in-situ of a chick forebrain by @danafd.bsky.social who studies early forebrain development in vertebrates & uses HCR to show the expression patterns of gene products instrumental in pallial/subpallial specification.
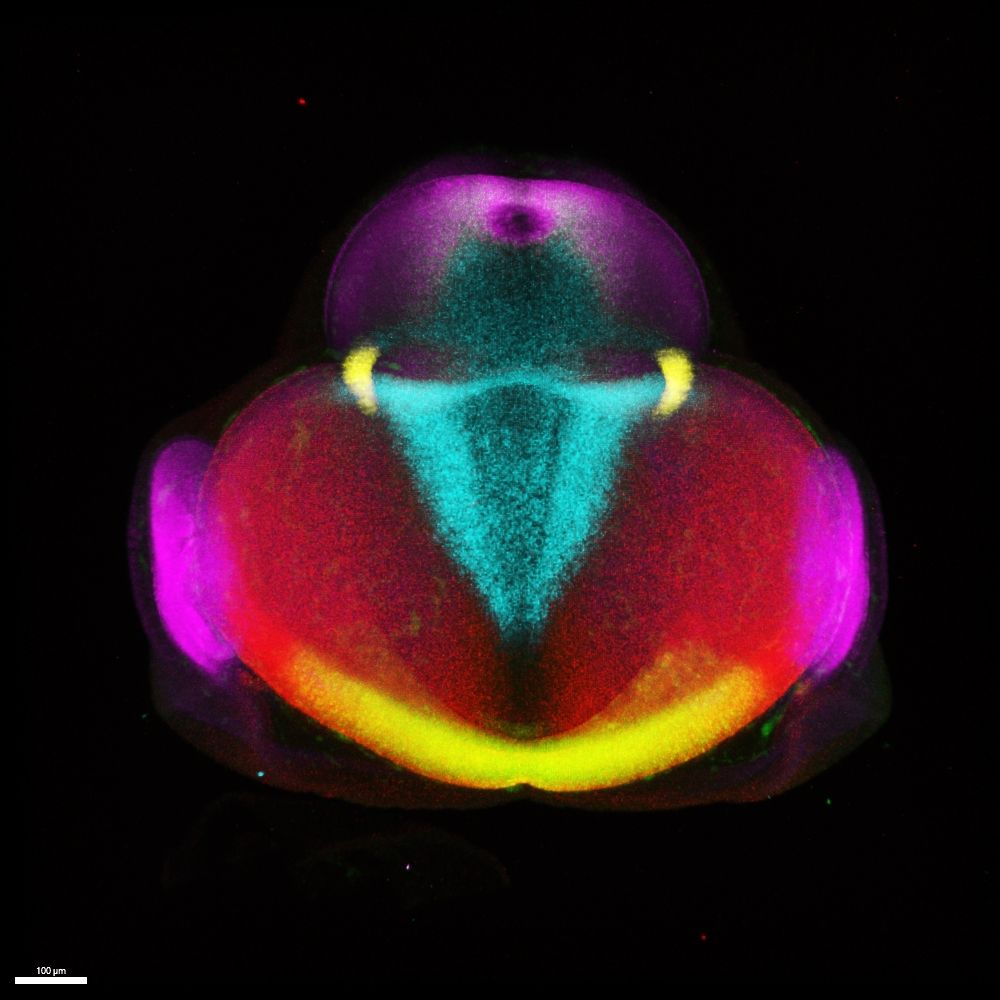
This week, it's a 3D image of a multiplex, in-situ of a chick forebrain by @danafd.bsky.social who studies early forebrain development in vertebrates & uses HCR to show the expression patterns of gene products instrumental in pallial/subpallial specification.
Here, José shows the building blocks of the developing cortex at 10pcw with SOX2 (apical progenitors), CTIP2 & TBR1(neuronal markers), NESTIN (radial glial fibers), and DAPI.
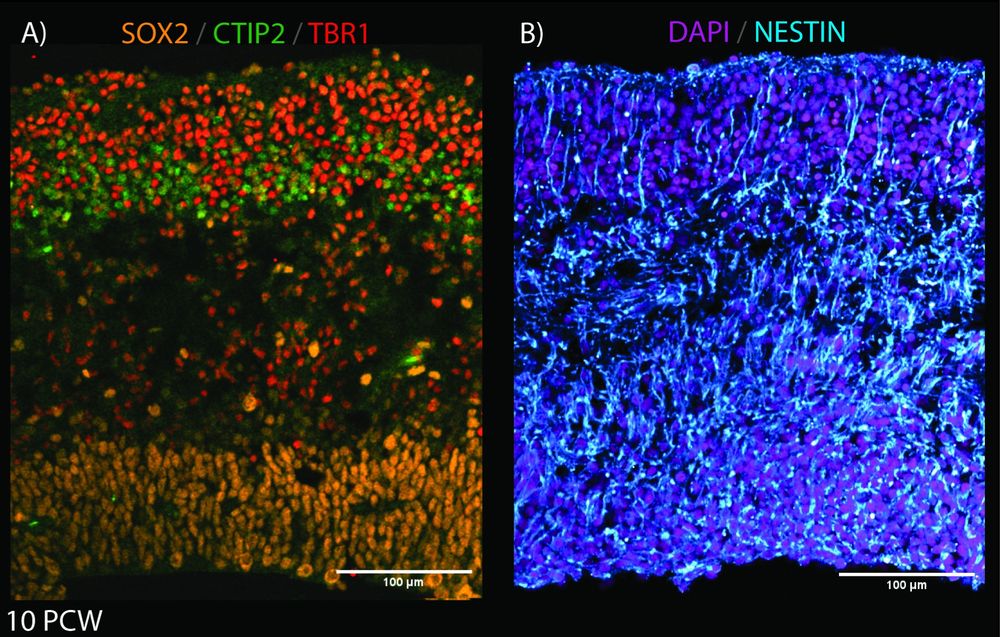
Here, José shows the building blocks of the developing cortex at 10pcw with SOX2 (apical progenitors), CTIP2 & TBR1(neuronal markers), NESTIN (radial glial fibers), and DAPI.
We had meaningful discussions and learned a lot from a vibrant community tackling tough questions on brain evolution and development! Super excited for what lies ahead!

We had meaningful discussions and learned a lot from a vibrant community tackling tough questions on brain evolution and development! Super excited for what lies ahead!

