
The Banfield Lab
@banfieldlab.bsky.social
Located at the University of California-Berkeley at the Innovative Genomics Institute.
Purveyors of Microbial Ecology, Bioinformatics, & Nanogeoscience.
Reposts or likes≠endorsements.
https://www.banfieldlab.com/
Purveyors of Microbial Ecology, Bioinformatics, & Nanogeoscience.
Reposts or likes≠endorsements.
https://www.banfieldlab.com/
Newest preprint from the group on megaplasmids in E. coli led by @assemblerag.bsky.social. In addition to our findings in metagenomic datasets, we identified an RNA-seq project from a giant panda gut sample with evidence of megaplasmids. We would like to include the data creators in our study.
October 6, 2025 at 11:42 PM
Newest preprint from the group on megaplasmids in E. coli led by @assemblerag.bsky.social. In addition to our findings in metagenomic datasets, we identified an RNA-seq project from a giant panda gut sample with evidence of megaplasmids. We would like to include the data creators in our study.
Reposted by The Banfield Lab
The 2025 Banfield Retreat was fantastic 🦠 so many talented scientists in one place is a sure way to come up with some great ideas!
@banfieldlab.bsky.social
@banfieldlab.bsky.social

May 1, 2025 at 6:06 PM
The 2025 Banfield Retreat was fantastic 🦠 so many talented scientists in one place is a sure way to come up with some great ideas!
@banfieldlab.bsky.social
@banfieldlab.bsky.social
Reposted by The Banfield Lab
🚨 Fresh from the press! We created and analyzed over 100 in vitro cyanobacterial consortia using well-characterized model cyanobacterial hosts to better understand how cyanobacteria recruit and interact with their microbiomes.
Check it out: doi.org/10.1093/isme...
Check it out: doi.org/10.1093/isme...

July 11, 2025 at 1:08 PM
🚨 Fresh from the press! We created and analyzed over 100 in vitro cyanobacterial consortia using well-characterized model cyanobacterial hosts to better understand how cyanobacteria recruit and interact with their microbiomes.
Check it out: doi.org/10.1093/isme...
Check it out: doi.org/10.1093/isme...
Reposted by The Banfield Lab
manuscript here! www.science.org/doi/10.1126/...

Overturning circulation structures the microbial functional seascape of the South Pacific
Global overturning circulation partitions the deep ocean into regions, each with different physicochemical characteristics, but the extent to which these water masses represent distinct ecosystems rem...
www.science.org
July 10, 2025 at 7:44 PM
manuscript here! www.science.org/doi/10.1126/...
Reposted by The Banfield Lab
Many of you gave me so much encouragement when I posted here earlier about my 10 year long passion project to make a microbial map of the ocean. Pinch me because I can't believe I get to update you that today that research was published in the journal SCIENCE! @science.org
July 10, 2025 at 7:29 PM
Many of you gave me so much encouragement when I posted here earlier about my 10 year long passion project to make a microbial map of the ocean. Pinch me because I can't believe I get to update you that today that research was published in the journal SCIENCE! @science.org
Reposted by The Banfield Lab
Out now:
Structure of an archaeal ribosome reveals a divergent active site and hibernation factor
@jhdcate.bsky.social @banfieldlab.bsky.social @diptinayak.bsky.social & co
#microsky
www.nature.com/articles/s41...
Structure of an archaeal ribosome reveals a divergent active site and hibernation factor
@jhdcate.bsky.social @banfieldlab.bsky.social @diptinayak.bsky.social & co
#microsky
www.nature.com/articles/s41...
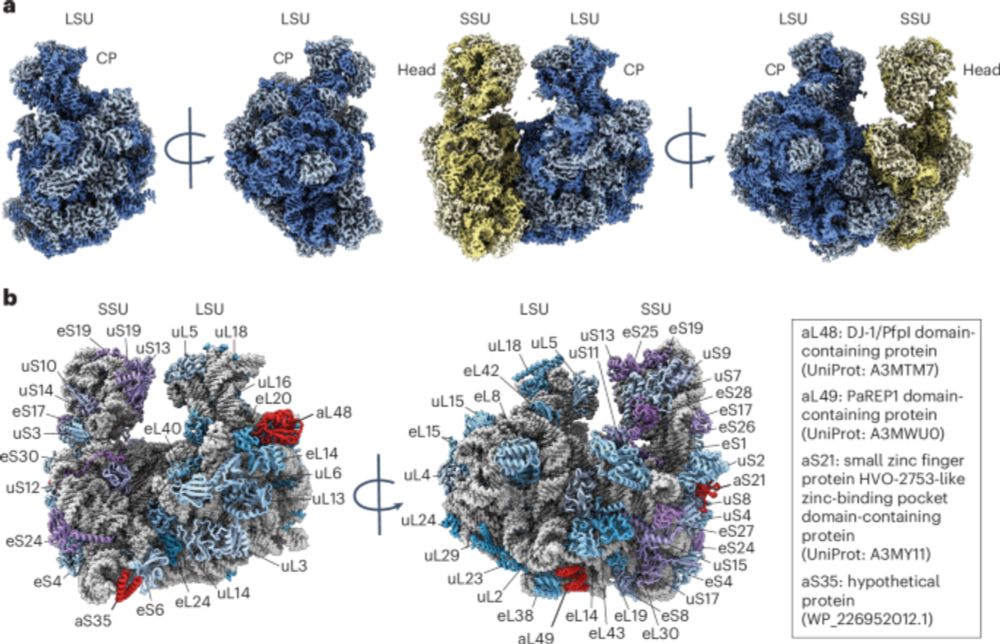
Structure of an archaeal ribosome reveals a divergent active site and hibernation factor - Nature Microbiology
Sequence and structural analyses reveal a divergent peptidyl transferase centre and a hibernation factor in archaea.
www.nature.com
July 18, 2025 at 11:02 AM
Out now:
Structure of an archaeal ribosome reveals a divergent active site and hibernation factor
@jhdcate.bsky.social @banfieldlab.bsky.social @diptinayak.bsky.social & co
#microsky
www.nature.com/articles/s41...
Structure of an archaeal ribosome reveals a divergent active site and hibernation factor
@jhdcate.bsky.social @banfieldlab.bsky.social @diptinayak.bsky.social & co
#microsky
www.nature.com/articles/s41...
Reposted by The Banfield Lab
In a recent paper, IGI's @jenniferdoudna.bsky.social @doudna-lab.bsky.social and Jill Banfield @banfieldlab.bsky.social collaborate with UCLA's @jacobsenucla.bsky.social to create a new, tiny #CRISPR tool for quicker, easier plant genome editing! Read more in this Q&A with Steve: ow.ly/6Szs50VGElf
Tiny CRISPR Tool Opens the Door to Faster, Simpler Plant Genome Editing
Tiny CRISPR Tool Opens the Door to Faster, Simpler Plant Genome Editing - Innovative Genomics Institute (IGI)
ow.ly
July 31, 2025 at 11:17 PM
In a recent paper, IGI's @jenniferdoudna.bsky.social @doudna-lab.bsky.social and Jill Banfield @banfieldlab.bsky.social collaborate with UCLA's @jacobsenucla.bsky.social to create a new, tiny #CRISPR tool for quicker, easier plant genome editing! Read more in this Q&A with Steve: ow.ly/6Szs50VGElf
Reposted by The Banfield Lab
Most plasmids described in E. coli are small compared to the megaplasmids we identified here! Check out the preprint if you want to learn about these mysterious large elements and their potential functions 🧬. I’m very grateful to have had the opportunity to work on this in @banfieldlab.bsky.social
Megaplasmids associate with Escherichia coli and other Enterobacteriaceae https://www.biorxiv.org/content/10.1101/2025.09.30.679422v1
October 1, 2025 at 5:10 AM
Most plasmids described in E. coli are small compared to the megaplasmids we identified here! Check out the preprint if you want to learn about these mysterious large elements and their potential functions 🧬. I’m very grateful to have had the opportunity to work on this in @banfieldlab.bsky.social
Excited to share new research led by our very own @lingdong-shi.bsky.social showing that many archaeal 23S rRNAs are circularized in active ribosomes. Check out the preprint below.
Happy to share our recent work on rRNA processing and conformation: www.biorxiv.org/content/10.1...
Huge thanks to @ppenev.bsky.social @Amos Nissley @Dipti Nayak @Rohan Sachdeva @jhdcate.bsky.social @Jill Banfield @banfieldlab.bsky.social !!
Huge thanks to @ppenev.bsky.social @Amos Nissley @Dipti Nayak @Rohan Sachdeva @jhdcate.bsky.social @Jill Banfield @banfieldlab.bsky.social !!

Circular 23S rRNA within archaeal ribosomes
The ribosomal RNAs (rRNAs) that form the core of ribosomes are believed to occur as linear molecules. Here, we investigated rRNAs from diverse and mostly uncultivated archaea and found evidence that, ...
www.biorxiv.org
April 29, 2025 at 9:34 PM
Excited to share new research led by our very own @lingdong-shi.bsky.social showing that many archaeal 23S rRNAs are circularized in active ribosomes. Check out the preprint below.
Genomes from long-read metagenomic assemblies contain rampant errors, highlighting the pressing need for stricter evaluation methods in long-read assembly algorithms. Read more in our paper with the Eren group. @floriantrigodet.bsky.social @merenbey.bsky.social
April 28, 2025 at 9:29 PM
Genomes from long-read metagenomic assemblies contain rampant errors, highlighting the pressing need for stricter evaluation methods in long-read assembly algorithms. Read more in our paper with the Eren group. @floriantrigodet.bsky.social @merenbey.bsky.social

