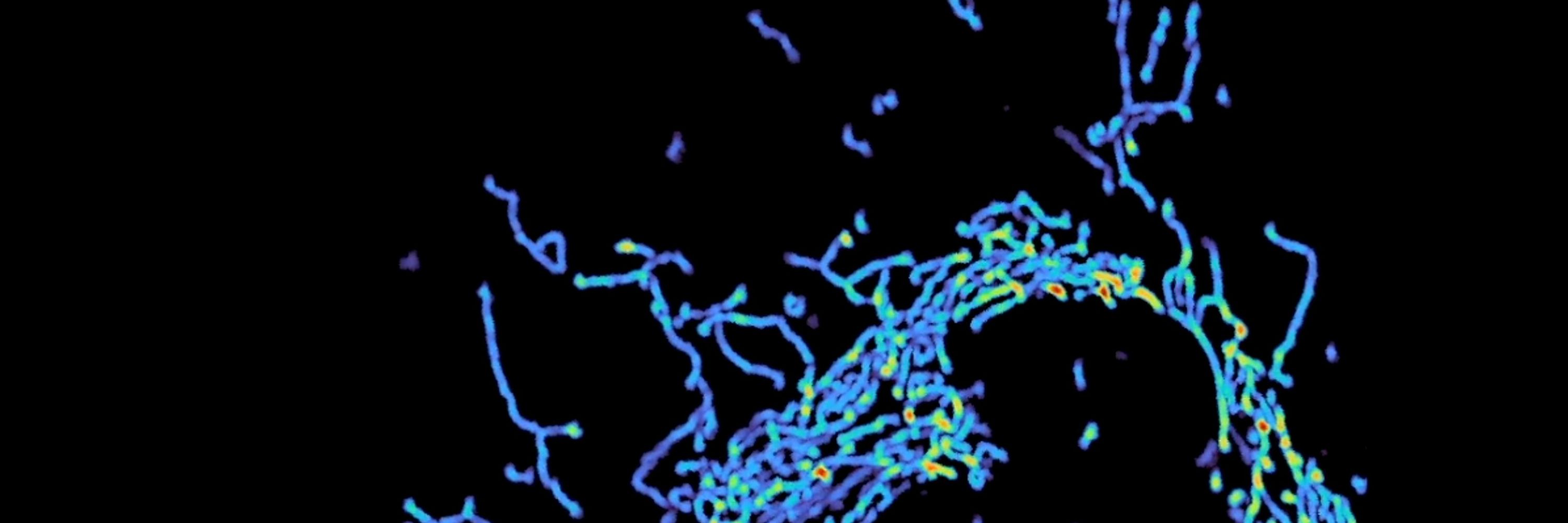
You can't spell imaging without "im aging" and boy am I.
Organelle enthusiast.
Creator of Nellie and Mitometer.
With an easy install and a click of a button, Nellie solves ALL these problems!
nature.com/articles/s41...
🧵2/N

With an easy install and a click of a button, Nellie solves ALL these problems!
nature.com/articles/s41...
🧵2/N
We've included some sample data if you want to explore what Nellie can do.
Code: github.com/aelefebv/nel...
Paper: nature.com/articles/s41...
And please share your results with us! We're excited to see how you'll use it in your research.
🧵Fin/N
We've included some sample data if you want to explore what Nellie can do.
Code: github.com/aelefebv/nel...
Paper: nature.com/articles/s41...
And please share your results with us! We're excited to see how you'll use it in your research.
🧵Fin/N
🧵14/N



🧵14/N
🧵13/N

🧵13/N
🧵12/N

🧵12/N
🧵11/N

🧵11/N
🧵10/N

🧵10/N
🧵9/N

🧵9/N
And of course @ritastrack.bsky.social for being a great editor!
🧵8/N
And of course @ritastrack.bsky.social for being a great editor!
🧵8/N
🧵7/N
🧵7/N
More fluorescence channels for your experiments!
🧵6/N
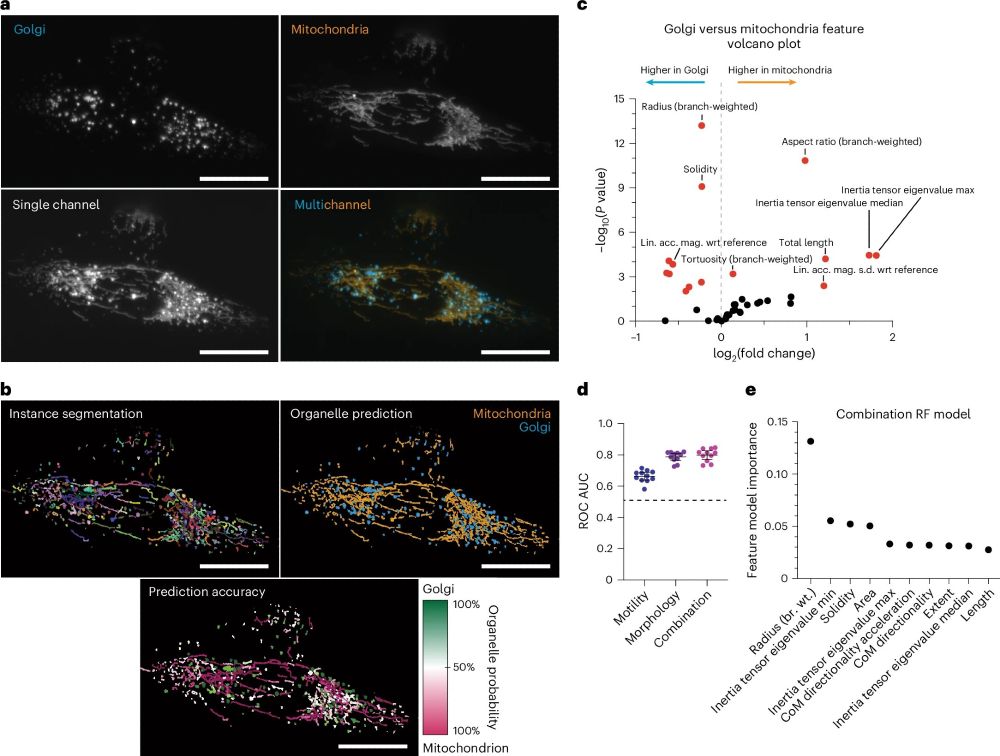
More fluorescence channels for your experiments!
🧵6/N

