You can't spell imaging without "im aging" and boy am I.
Organelle enthusiast.
Creator of Nellie and Mitometer.
🧵14/N



🧵14/N
🧵13/N

🧵13/N
🧵12/N

🧵12/N
🧵11/N

🧵11/N
🧵10/N

🧵10/N
🧵9/N

🧵9/N
🧵7/N
🧵7/N
More fluorescence channels for your experiments!
🧵6/N
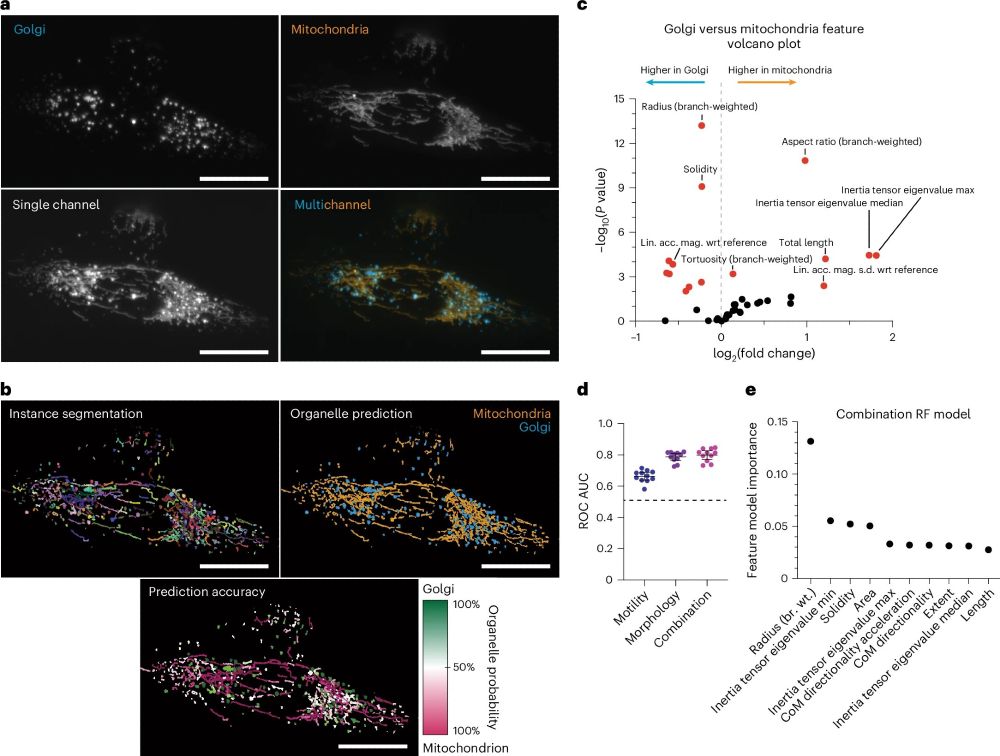
More fluorescence channels for your experiments!
🧵6/N
🧵5/N

🧵5/N
🧵4/N

🧵4/N
(credit to @andrewgyork.bsky.social and @amsikking.bsky.social 's Snouty SOLS microscope for the data, and @alisterburt.bsky.social for the Animation plugin)
🧵3/N
(credit to @andrewgyork.bsky.social and @amsikking.bsky.social 's Snouty SOLS microscope for the data, and @alisterburt.bsky.social for the Animation plugin)
🧵3/N
Well I am THRILLED to announce that Nellie has been published in @naturemethods.bsky.social!
Nellie a fully automated pipeline for organelle segmentation, tracking, and hierarchical feature extraction in 2D, 3D, timelapse, multichannel live-cell microscopy
🧵1/N
Well I am THRILLED to announce that Nellie has been published in @naturemethods.bsky.social!
Nellie a fully automated pipeline for organelle segmentation, tracking, and hierarchical feature extraction in 2D, 3D, timelapse, multichannel live-cell microscopy
🧵1/N

