
Arc Institute
@arcinstitute.org
A new scientific institution for curiosity-driven biomedical science and technology.
Hear from lead author, Alison Fanton as she walks through how the team achieved these results and what they could mean for the future of gene editing.
Blog: arcinstitute.org/news/large-s...
Video: youtube.com/watch?time_continue=1&v=DHg_TkN15XY
Blog: arcinstitute.org/news/large-s...
Video: youtube.com/watch?time_continue=1&v=DHg_TkN15XY

Site-specific DNA insertion into the human genome with engineered recombinases
YouTube video by Arc Institute
youtube.com
November 6, 2025 at 5:46 PM
Hear from lead author, Alison Fanton as she walks through how the team achieved these results and what they could mean for the future of gene editing.
Blog: arcinstitute.org/news/large-s...
Video: youtube.com/watch?time_continue=1&v=DHg_TkN15XY
Blog: arcinstitute.org/news/large-s...
Video: youtube.com/watch?time_continue=1&v=DHg_TkN15XY
The optimized enzymes work across difficult cell types, with 33% efficiency in non-dividing cells, 24% in embryonic stem cells & 17% in human T cells, providing a new blueprint for engineering precise genome-editing tools.
Learn more in the paper: www.nature.com/articles/s41...
Learn more in the paper: www.nature.com/articles/s41...

Site-specific DNA insertion into the human genome with engineered recombinases - Nature Biotechnology
Engineered DNA recombinases efficiently and specifically insert genetic cargos without the use of landing pads.
www.nature.com
November 6, 2025 at 5:46 PM
The optimized enzymes work across difficult cell types, with 33% efficiency in non-dividing cells, 24% in embryonic stem cells & 17% in human T cells, providing a new blueprint for engineering precise genome-editing tools.
Learn more in the paper: www.nature.com/articles/s41...
Learn more in the paper: www.nature.com/articles/s41...
The best variants achieved 97% specificity & up to 53% efficiency, a 7.5-fold increase in accuracy & 12-fold increase in efficiency over the starting enzyme.
This means researchers can now choose variants optimized for maximum efficiency, specificity, or a balance of both.
This means researchers can now choose variants optimized for maximum efficiency, specificity, or a balance of both.

November 6, 2025 at 5:46 PM
The best variants achieved 97% specificity & up to 53% efficiency, a 7.5-fold increase in accuracy & 12-fold increase in efficiency over the starting enzyme.
This means researchers can now choose variants optimized for maximum efficiency, specificity, or a balance of both.
This means researchers can now choose variants optimized for maximum efficiency, specificity, or a balance of both.
They tested thousands of mutations to identify which improved the enzyme, then used computational models to predict how combining mutations would impact performance, allowing them to build highly optimized variants rapidly.

November 6, 2025 at 5:46 PM
They tested thousands of mutations to identify which improved the enzyme, then used computational models to predict how combining mutations would impact performance, allowing them to build highly optimized variants rapidly.
The team developed a comprehensive engineering strategy to improve both efficiency & specificity, combining evolutionary screening to find better mutations, machine learning to predict which mutations work together & fusing the enzyme to dCas9 to guide it to the correct location.

November 6, 2025 at 5:46 PM
The team developed a comprehensive engineering strategy to improve both efficiency & specificity, combining evolutionary screening to find better mutations, machine learning to predict which mutations work together & fusing the enzyme to dCas9 to guide it to the correct location.
Recombinases are enzymes capable of inserting DNA at specific sites in the genome without needing to create double-strand breaks like CRISPR does.
Existing recombinases, however, have limitations–managing only ~5% efficiency & often hitting hundreds of off-target sites.
Existing recombinases, however, have limitations–managing only ~5% efficiency & often hitting hundreds of off-target sites.

November 6, 2025 at 5:46 PM
Recombinases are enzymes capable of inserting DNA at specific sites in the genome without needing to create double-strand breaks like CRISPR does.
Existing recombinases, however, have limitations–managing only ~5% efficiency & often hitting hundreds of off-target sites.
Existing recombinases, however, have limitations–managing only ~5% efficiency & often hitting hundreds of off-target sites.
By showing PTGES3 is an essential AR regulator, this work points to a new therapeutic strategy for overcoming treatment resistance in metastatic prostate cancer.
Learn more in the full paper: www.nature.com/articles/s41...
Learn more in the full paper: www.nature.com/articles/s41...

Genome-scale CRISPR screens identify PTGES3 as a direct modulator of androgen receptor function in advanced prostate cancer - Nature Genetics
Genome-wide CRISPRi screens for modulators of androgen receptor (AR) protein levels using live-cell quantitative endogenous AR fluorescence reporters identify PTGES3 as a new regulator of AR stability...
www.nature.com
November 5, 2025 at 4:21 PM
By showing PTGES3 is an essential AR regulator, this work points to a new therapeutic strategy for overcoming treatment resistance in metastatic prostate cancer.
Learn more in the full paper: www.nature.com/articles/s41...
Learn more in the full paper: www.nature.com/articles/s41...
Importantly, PTGES3 is selectively required in AR-driven prostate cancer cells but not AR-independent cancer cells.
Unlike targeting HSP90, inhibiting PTGES3 offers a potentially more specific way to block AR without broadly disrupting the heat shock response.
Unlike targeting HSP90, inhibiting PTGES3 offers a potentially more specific way to block AR without broadly disrupting the heat shock response.

November 5, 2025 at 4:21 PM
Importantly, PTGES3 is selectively required in AR-driven prostate cancer cells but not AR-independent cancer cells.
Unlike targeting HSP90, inhibiting PTGES3 offers a potentially more specific way to block AR without broadly disrupting the heat shock response.
Unlike targeting HSP90, inhibiting PTGES3 offers a potentially more specific way to block AR without broadly disrupting the heat shock response.
In clinical findings, high PTGES3 expression predicts worse outcomes in patients treated with drugs like abiraterone & enzalutamide.
But loss of PTGES3 blocked tumor growth across multiple models of aggressive, therapy-resistant prostate cancer.
But loss of PTGES3 blocked tumor growth across multiple models of aggressive, therapy-resistant prostate cancer.

November 5, 2025 at 4:21 PM
In clinical findings, high PTGES3 expression predicts worse outcomes in patients treated with drugs like abiraterone & enzalutamide.
But loss of PTGES3 blocked tumor growth across multiple models of aggressive, therapy-resistant prostate cancer.
But loss of PTGES3 blocked tumor growth across multiple models of aggressive, therapy-resistant prostate cancer.
Through biochemical experiments & structural modeling, researchers showed that PTGES3 binds directly to AR in the nucleus. This binding interaction stabilizes AR protein & is required for AR to activate its target genes.

November 5, 2025 at 4:21 PM
Through biochemical experiments & structural modeling, researchers showed that PTGES3 binds directly to AR in the nucleus. This binding interaction stabilizes AR protein & is required for AR to activate its target genes.
The team developed an endogenous AR fluorescent reporter & used genome-wide CRISPRi screens to systematically map genes that control AR protein abundance.
PTGES3 was a top hit, but almost nothing was known about its role in prostate cancer.
PTGES3 was a top hit, but almost nothing was known about its role in prostate cancer.

November 5, 2025 at 4:21 PM
The team developed an endogenous AR fluorescent reporter & used genome-wide CRISPRi screens to systematically map genes that control AR protein abundance.
PTGES3 was a top hit, but almost nothing was known about its role in prostate cancer.
PTGES3 was a top hit, but almost nothing was known about its role in prostate cancer.
The Pluvinage Lab focuses on identifying pathogenic autoantibodies in neurological diseases and designing antigen-specific therapies to eliminate them. Learn more: arcinstitute.org/labs/pluvina...

Pluvinage Lab - Arc Institute
Arc Institute is a independent nonprofit research organization headquartered in Palo Alto, California.
arcinstitute.org
October 29, 2025 at 3:25 PM
The Pluvinage Lab focuses on identifying pathogenic autoantibodies in neurological diseases and designing antigen-specific therapies to eliminate them. Learn more: arcinstitute.org/labs/pluvina...
Drawing from clinical neurology training and his discovery of a previously undetectable form of B12 deficiency affecting dementia patients, he brings a unique perspective bridging the lab and clinic to Arc: arcinstitute.org/news/meet-jo...

Meet John Pluvinage, Arc’s 9th Core Investigator and first physician-scientist | Arc Institute
John Pluvinage (X: @jvpluv), Arc’s 9th Core Investigator and first physician-scientist, has arrived and is now setting up his lab. Pluvinage investigates the overlap between autoimmunity and neurodege...
arcinstitute.org
October 29, 2025 at 3:25 PM
Drawing from clinical neurology training and his discovery of a previously undetectable form of B12 deficiency affecting dementia patients, he brings a unique perspective bridging the lab and clinic to Arc: arcinstitute.org/news/meet-jo...
For a deeper look at the models, challenges, and future directions, read our conversation with project leads
@genophoria.bsky.social, Core Investigator at Arc & Laksshman Sundaram, Director of Applied Research at
NVIDIA: arcinstitute.org/news/codonfm...
@genophoria.bsky.social, Core Investigator at Arc & Laksshman Sundaram, Director of Applied Research at
NVIDIA: arcinstitute.org/news/codonfm...

The Hidden “Grammar” Revealed by CodonFM, our Foundation Models for Codons | Arc Institute
A collaboration between Arc Institute and NVIDIA has now developed CodonFM, a family of open source AI models that reveal the intricate grammar underlying codon choice. The work demonstrates that codo...
arcinstitute.org
October 28, 2025 at 8:55 PM
For a deeper look at the models, challenges, and future directions, read our conversation with project leads
@genophoria.bsky.social, Core Investigator at Arc & Laksshman Sundaram, Director of Applied Research at
NVIDIA: arcinstitute.org/news/codonfm...
@genophoria.bsky.social, Core Investigator at Arc & Laksshman Sundaram, Director of Applied Research at
NVIDIA: arcinstitute.org/news/codonfm...
By modeling codon usage directly, CodonFM achieves multi-fold improvements over prior approaches, revealing how synonymous variation influences expression, variant impact, and therapeutic design.
Learn more in the preprint: research.nvidia.com/labs/dbr/ass...
Learn more in the preprint: research.nvidia.com/labs/dbr/ass...

October 28, 2025 at 8:55 PM
By modeling codon usage directly, CodonFM achieves multi-fold improvements over prior approaches, revealing how synonymous variation influences expression, variant impact, and therapeutic design.
Learn more in the preprint: research.nvidia.com/labs/dbr/ass...
Learn more in the preprint: research.nvidia.com/labs/dbr/ass...
As the model scaled, clear grammar began to emerge.
Smaller versions hinted at patterns, but at the billion-parameter scale, CodonFM could predict which codons cells would choose in context––revealing long-range dependencies that link codon choice to translation and expression.
Smaller versions hinted at patterns, but at the billion-parameter scale, CodonFM could predict which codons cells would choose in context––revealing long-range dependencies that link codon choice to translation and expression.

October 28, 2025 at 8:55 PM
As the model scaled, clear grammar began to emerge.
Smaller versions hinted at patterns, but at the billion-parameter scale, CodonFM could predict which codons cells would choose in context––revealing long-range dependencies that link codon choice to translation and expression.
Smaller versions hinted at patterns, but at the billion-parameter scale, CodonFM could predict which codons cells would choose in context––revealing long-range dependencies that link codon choice to translation and expression.
Two complementary architectures power the CodonFM family.
Encodon, released today, interprets codon context to understand regulatory effects and mutations, and Decodon, to be released later this year, generates optimized sequences for design applications.
Encodon, released today, interprets codon context to understand regulatory effects and mutations, and Decodon, to be released later this year, generates optimized sequences for design applications.

October 28, 2025 at 8:55 PM
Two complementary architectures power the CodonFM family.
Encodon, released today, interprets codon context to understand regulatory effects and mutations, and Decodon, to be released later this year, generates optimized sequences for design applications.
Encodon, released today, interprets codon context to understand regulatory effects and mutations, and Decodon, to be released later this year, generates optimized sequences for design applications.
Trained on 130 million coding sequences from more than 20,000 species, CodonFM uses large-scale language modeling to uncover the patterns behind codon choice and reveal the regulatory logic that links sequence variation to gene expression and protein abundance.

October 28, 2025 at 8:55 PM
Trained on 130 million coding sequences from more than 20,000 species, CodonFM uses large-scale language modeling to uncover the patterns behind codon choice and reveal the regulatory logic that links sequence variation to gene expression and protein abundance.
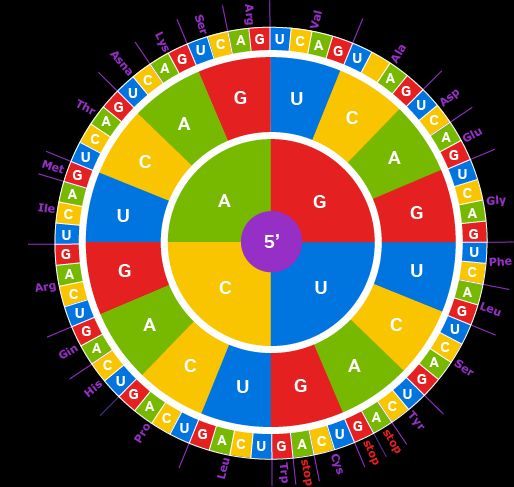
There is a natural redundancy in the genetic code. 64 codons encode 20 amino acids, resulting in multiple "synonymous" codons encoding for the same amino acid.
Some of these, however, appear far more often than others, following consistent, non-random patterns across genes.
Some of these, however, appear far more often than others, following consistent, non-random patterns across genes.
October 28, 2025 at 8:55 PM
There is a natural redundancy in the genetic code. 64 codons encode 20 amino acids, resulting in multiple "synonymous" codons encoding for the same amino acid.
Some of these, however, appear far more often than others, following consistent, non-random patterns across genes.
Some of these, however, appear far more often than others, following consistent, non-random patterns across genes.
By demonstrating that human T cells can be reprogrammed without the need to cut DNA, this work offers a path towards safer, more effective treatments across cancer, transplant medicine, and autoimmune disease.
Learn more in the full paper: www.nature.com/articles/s41...
Learn more in the full paper: www.nature.com/articles/s41...

Integrated epigenetic and genetic programming of primary human T cells - Nature Biotechnology
Multiplexed editing in primary human T cells generates enhanced immune cell therapies.
www.nature.com
October 21, 2025 at 4:43 PM
By demonstrating that human T cells can be reprogrammed without the need to cut DNA, this work offers a path towards safer, more effective treatments across cancer, transplant medicine, and autoimmune disease.
Learn more in the full paper: www.nature.com/articles/s41...
Learn more in the full paper: www.nature.com/articles/s41...

