
Arc Institute
@arcinstitute.org
A new scientific institution for curiosity-driven biomedical science and technology.
The best variants achieved 97% specificity & up to 53% efficiency, a 7.5-fold increase in accuracy & 12-fold increase in efficiency over the starting enzyme.
This means researchers can now choose variants optimized for maximum efficiency, specificity, or a balance of both.
This means researchers can now choose variants optimized for maximum efficiency, specificity, or a balance of both.

November 6, 2025 at 5:46 PM
The best variants achieved 97% specificity & up to 53% efficiency, a 7.5-fold increase in accuracy & 12-fold increase in efficiency over the starting enzyme.
This means researchers can now choose variants optimized for maximum efficiency, specificity, or a balance of both.
This means researchers can now choose variants optimized for maximum efficiency, specificity, or a balance of both.
They tested thousands of mutations to identify which improved the enzyme, then used computational models to predict how combining mutations would impact performance, allowing them to build highly optimized variants rapidly.

November 6, 2025 at 5:46 PM
They tested thousands of mutations to identify which improved the enzyme, then used computational models to predict how combining mutations would impact performance, allowing them to build highly optimized variants rapidly.
The team developed a comprehensive engineering strategy to improve both efficiency & specificity, combining evolutionary screening to find better mutations, machine learning to predict which mutations work together & fusing the enzyme to dCas9 to guide it to the correct location.

November 6, 2025 at 5:46 PM
The team developed a comprehensive engineering strategy to improve both efficiency & specificity, combining evolutionary screening to find better mutations, machine learning to predict which mutations work together & fusing the enzyme to dCas9 to guide it to the correct location.
Recombinases are enzymes capable of inserting DNA at specific sites in the genome without needing to create double-strand breaks like CRISPR does.
Existing recombinases, however, have limitations–managing only ~5% efficiency & often hitting hundreds of off-target sites.
Existing recombinases, however, have limitations–managing only ~5% efficiency & often hitting hundreds of off-target sites.

November 6, 2025 at 5:46 PM
Recombinases are enzymes capable of inserting DNA at specific sites in the genome without needing to create double-strand breaks like CRISPR does.
Existing recombinases, however, have limitations–managing only ~5% efficiency & often hitting hundreds of off-target sites.
Existing recombinases, however, have limitations–managing only ~5% efficiency & often hitting hundreds of off-target sites.
Work published today in @natbiotech.nature.com from Arc’s Luke Gilbert and Patrick Hsu labs presents a new way to insert large DNA sequences into the genome using engineered recombinases that don’t require DNA cutting or rely on the cell's repair machinery.

November 6, 2025 at 5:46 PM
Work published today in @natbiotech.nature.com from Arc’s Luke Gilbert and Patrick Hsu labs presents a new way to insert large DNA sequences into the genome using engineered recombinases that don’t require DNA cutting or rely on the cell's repair machinery.
Importantly, PTGES3 is selectively required in AR-driven prostate cancer cells but not AR-independent cancer cells.
Unlike targeting HSP90, inhibiting PTGES3 offers a potentially more specific way to block AR without broadly disrupting the heat shock response.
Unlike targeting HSP90, inhibiting PTGES3 offers a potentially more specific way to block AR without broadly disrupting the heat shock response.

November 5, 2025 at 4:21 PM
Importantly, PTGES3 is selectively required in AR-driven prostate cancer cells but not AR-independent cancer cells.
Unlike targeting HSP90, inhibiting PTGES3 offers a potentially more specific way to block AR without broadly disrupting the heat shock response.
Unlike targeting HSP90, inhibiting PTGES3 offers a potentially more specific way to block AR without broadly disrupting the heat shock response.
In clinical findings, high PTGES3 expression predicts worse outcomes in patients treated with drugs like abiraterone & enzalutamide.
But loss of PTGES3 blocked tumor growth across multiple models of aggressive, therapy-resistant prostate cancer.
But loss of PTGES3 blocked tumor growth across multiple models of aggressive, therapy-resistant prostate cancer.

November 5, 2025 at 4:21 PM
In clinical findings, high PTGES3 expression predicts worse outcomes in patients treated with drugs like abiraterone & enzalutamide.
But loss of PTGES3 blocked tumor growth across multiple models of aggressive, therapy-resistant prostate cancer.
But loss of PTGES3 blocked tumor growth across multiple models of aggressive, therapy-resistant prostate cancer.
Through biochemical experiments & structural modeling, researchers showed that PTGES3 binds directly to AR in the nucleus. This binding interaction stabilizes AR protein & is required for AR to activate its target genes.

November 5, 2025 at 4:21 PM
Through biochemical experiments & structural modeling, researchers showed that PTGES3 binds directly to AR in the nucleus. This binding interaction stabilizes AR protein & is required for AR to activate its target genes.
The team developed an endogenous AR fluorescent reporter & used genome-wide CRISPRi screens to systematically map genes that control AR protein abundance.
PTGES3 was a top hit, but almost nothing was known about its role in prostate cancer.
PTGES3 was a top hit, but almost nothing was known about its role in prostate cancer.

November 5, 2025 at 4:21 PM
The team developed an endogenous AR fluorescent reporter & used genome-wide CRISPRi screens to systematically map genes that control AR protein abundance.
PTGES3 was a top hit, but almost nothing was known about its role in prostate cancer.
PTGES3 was a top hit, but almost nothing was known about its role in prostate cancer.
A study out today in @natgenet.nature.com from Core Investigator Luke Gilbert, Haolong Li, & others identifies PTGES3 as a critical regulator of androgen receptor (AR) protein levels in prostate cancer, revealing a potential new target for tumors that become treatment-resistant.

November 5, 2025 at 4:21 PM
A study out today in @natgenet.nature.com from Core Investigator Luke Gilbert, Haolong Li, & others identifies PTGES3 as a critical regulator of androgen receptor (AR) protein levels in prostate cancer, revealing a potential new target for tumors that become treatment-resistant.
Welcome to our 9th Core Investigator and first physician-scientist, John Pluvinage. His team investigates the hidden overlap between autoimmunity and neurodegeneration, developing targeted treatments for mysterious neurological cases and common dementias.

October 29, 2025 at 3:25 PM
Welcome to our 9th Core Investigator and first physician-scientist, John Pluvinage. His team investigates the hidden overlap between autoimmunity and neurodegeneration, developing targeted treatments for mysterious neurological cases and common dementias.
By modeling codon usage directly, CodonFM achieves multi-fold improvements over prior approaches, revealing how synonymous variation influences expression, variant impact, and therapeutic design.
Learn more in the preprint: research.nvidia.com/labs/dbr/ass...
Learn more in the preprint: research.nvidia.com/labs/dbr/ass...

October 28, 2025 at 8:55 PM
By modeling codon usage directly, CodonFM achieves multi-fold improvements over prior approaches, revealing how synonymous variation influences expression, variant impact, and therapeutic design.
Learn more in the preprint: research.nvidia.com/labs/dbr/ass...
Learn more in the preprint: research.nvidia.com/labs/dbr/ass...
As the model scaled, clear grammar began to emerge.
Smaller versions hinted at patterns, but at the billion-parameter scale, CodonFM could predict which codons cells would choose in context––revealing long-range dependencies that link codon choice to translation and expression.
Smaller versions hinted at patterns, but at the billion-parameter scale, CodonFM could predict which codons cells would choose in context––revealing long-range dependencies that link codon choice to translation and expression.

October 28, 2025 at 8:55 PM
As the model scaled, clear grammar began to emerge.
Smaller versions hinted at patterns, but at the billion-parameter scale, CodonFM could predict which codons cells would choose in context––revealing long-range dependencies that link codon choice to translation and expression.
Smaller versions hinted at patterns, but at the billion-parameter scale, CodonFM could predict which codons cells would choose in context––revealing long-range dependencies that link codon choice to translation and expression.
Two complementary architectures power the CodonFM family.
Encodon, released today, interprets codon context to understand regulatory effects and mutations, and Decodon, to be released later this year, generates optimized sequences for design applications.
Encodon, released today, interprets codon context to understand regulatory effects and mutations, and Decodon, to be released later this year, generates optimized sequences for design applications.

October 28, 2025 at 8:55 PM
Two complementary architectures power the CodonFM family.
Encodon, released today, interprets codon context to understand regulatory effects and mutations, and Decodon, to be released later this year, generates optimized sequences for design applications.
Encodon, released today, interprets codon context to understand regulatory effects and mutations, and Decodon, to be released later this year, generates optimized sequences for design applications.
Trained on 130 million coding sequences from more than 20,000 species, CodonFM uses large-scale language modeling to uncover the patterns behind codon choice and reveal the regulatory logic that links sequence variation to gene expression and protein abundance.

October 28, 2025 at 8:55 PM
Trained on 130 million coding sequences from more than 20,000 species, CodonFM uses large-scale language modeling to uncover the patterns behind codon choice and reveal the regulatory logic that links sequence variation to gene expression and protein abundance.
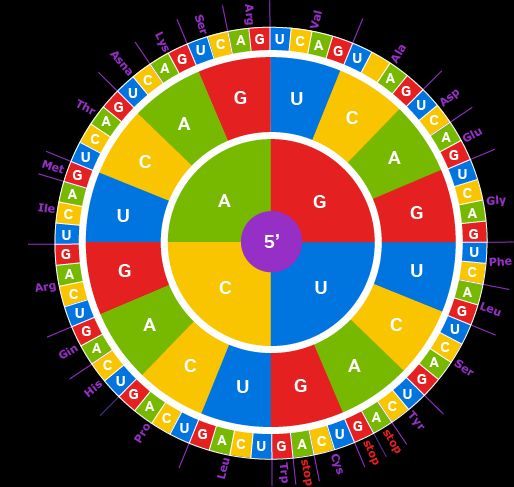
There is a natural redundancy in the genetic code. 64 codons encode 20 amino acids, resulting in multiple "synonymous" codons encoding for the same amino acid.
Some of these, however, appear far more often than others, following consistent, non-random patterns across genes.
Some of these, however, appear far more often than others, following consistent, non-random patterns across genes.
October 28, 2025 at 8:55 PM
There is a natural redundancy in the genetic code. 64 codons encode 20 amino acids, resulting in multiple "synonymous" codons encoding for the same amino acid.
Some of these, however, appear far more often than others, following consistent, non-random patterns across genes.
Some of these, however, appear far more often than others, following consistent, non-random patterns across genes.
Arc Core Investigator @genophoria.bsky.social will speak on a free, online panel with @tkaraletsos.bsky.social, @emmalundberg.bsky.social, and @ronalfa.bsky.social on Wed., Oct. 29, as part of GENbio's “The State of AI in Drug Discovery in 2025.” Register at: webinars.liebertpub.com/e/the-state-...

October 24, 2025 at 5:32 PM
Arc Core Investigator @genophoria.bsky.social will speak on a free, online panel with @tkaraletsos.bsky.social, @emmalundberg.bsky.social, and @ronalfa.bsky.social on Wed., Oct. 29, as part of GENbio's “The State of AI in Drug Discovery in 2025.” Register at: webinars.liebertpub.com/e/the-state-...
To test the approach's durability, the team monitored the protein product for each of the targeted genes over a period of 28 days.
Expression remained stable for up to 80 generations, showing that CRISPRoff gene silencing produces highly specific and sustained modifications.
Expression remained stable for up to 80 generations, showing that CRISPRoff gene silencing produces highly specific and sustained modifications.

October 21, 2025 at 4:43 PM
To test the approach's durability, the team monitored the protein product for each of the targeted genes over a period of 28 days.
Expression remained stable for up to 80 generations, showing that CRISPRoff gene silencing produces highly specific and sustained modifications.
Expression remained stable for up to 80 generations, showing that CRISPRoff gene silencing produces highly specific and sustained modifications.
Unlike traditional CRISPR approaches requiring double-strand DNA breaks that can harm or kill T cells, the team shows that CRISPRon & CRISPRoff are capable of modifying up to five genes simultaneously while maintaining high rates of cell survival.

October 21, 2025 at 4:43 PM
Unlike traditional CRISPR approaches requiring double-strand DNA breaks that can harm or kill T cells, the team shows that CRISPRon & CRISPRoff are capable of modifying up to five genes simultaneously while maintaining high rates of cell survival.
The all-RNA platform, composed of CRISPRoff & CRISPRon epigenetic editors, allows the team to stably modify gene expression by adding or removing methylation marks that can silence or activate genes.

October 21, 2025 at 4:43 PM
The all-RNA platform, composed of CRISPRoff & CRISPRon epigenetic editors, allows the team to stably modify gene expression by adding or removing methylation marks that can silence or activate genes.
Published today in @natureportfolio.nature.com Biotechnology, Laine Goudy, Luke Gilbert, Alex Marson, & colleagues report an epigenetic editing platform that safely reprograms multiple genes in human T cells without many of the challenges & risks associated with traditional gene editing approaches.

October 21, 2025 at 4:43 PM
Published today in @natureportfolio.nature.com Biotechnology, Laine Goudy, Luke Gilbert, Alex Marson, & colleagues report an epigenetic editing platform that safely reprograms multiple genes in human T cells without many of the challenges & risks associated with traditional gene editing approaches.
Arc is looking for an outstanding early-career scientist to be our next Science Fellow. This program provides resources and freedom for those looking to transition into a principal investigator role immediately after doctoral training. Apply here: arcinstitute.org/programs/sea...

October 13, 2025 at 7:36 PM
Arc is looking for an outstanding early-career scientist to be our next Science Fellow. This program provides resources and freedom for those looking to transition into a principal investigator role immediately after doctoral training. Apply here: arcinstitute.org/programs/sea...
Hear how Arc, Ultima Genomics, and @10xgenomics.bsky.social are partnering to generate perturbation data at the scale needed to train virtual cell models on The Bio Report podcast with guests @genophoria.bsky.social, Gilad Almogy, and
Serge Saxonov: thebioreport.podbean.com/e/transformi...
Serge Saxonov: thebioreport.podbean.com/e/transformi...

October 9, 2025 at 4:51 PM
Hear how Arc, Ultima Genomics, and @10xgenomics.bsky.social are partnering to generate perturbation data at the scale needed to train virtual cell models on The Bio Report podcast with guests @genophoria.bsky.social, Gilad Almogy, and
Serge Saxonov: thebioreport.podbean.com/e/transformi...
Serge Saxonov: thebioreport.podbean.com/e/transformi...
The optimized ISCro4 system was then tested in proof-of-concept edits – including excision of Friedreich’s ataxia–associated repeats and deletion of BCL11A , a validated sickle cell target – suggesting the technology's potential for therapeutic application.

September 25, 2025 at 6:27 PM
The optimized ISCro4 system was then tested in proof-of-concept edits – including excision of Friedreich’s ataxia–associated repeats and deletion of BCL11A , a validated sickle cell target – suggesting the technology's potential for therapeutic application.
Through systematic engineering of both the recombinase protein and its guide RNA guide, the researchers optimized ISCro4, reaching up to 20% insertion efficiency and 82% specificity for hitting its intended targets in the human genome.

September 25, 2025 at 6:27 PM
Through systematic engineering of both the recombinase protein and its guide RNA guide, the researchers optimized ISCro4, reaching up to 20% insertion efficiency and 82% specificity for hitting its intended targets in the human genome.

