
Shiny app to explore results in manuscript: connect.doit.wisc.edu/content/122/

Shiny app to explore results in manuscript: connect.doit.wisc.edu/content/122/
- data-driven omic data discretization
- permutation testing to eliminate spurious predictions
- full workflow and downstream analyses in an R package
- Shiny app for interactive visualization
- data-driven omic data discretization
- permutation testing to eliminate spurious predictions
- full workflow and downstream analyses in an R package
- Shiny app for interactive visualization
I may test how many calls are possible with the free academic plan to see if it is worthwhile to update my repo.

I may test how many calls are possible with the free academic plan to see if it is worthwhile to update my repo.





GitHub: github.com/gitter-lab/A...
Datasets: doi.org/10.5281/zeno...
7/

GitHub: github.com/gitter-lab/A...
Datasets: doi.org/10.5281/zeno...
7/
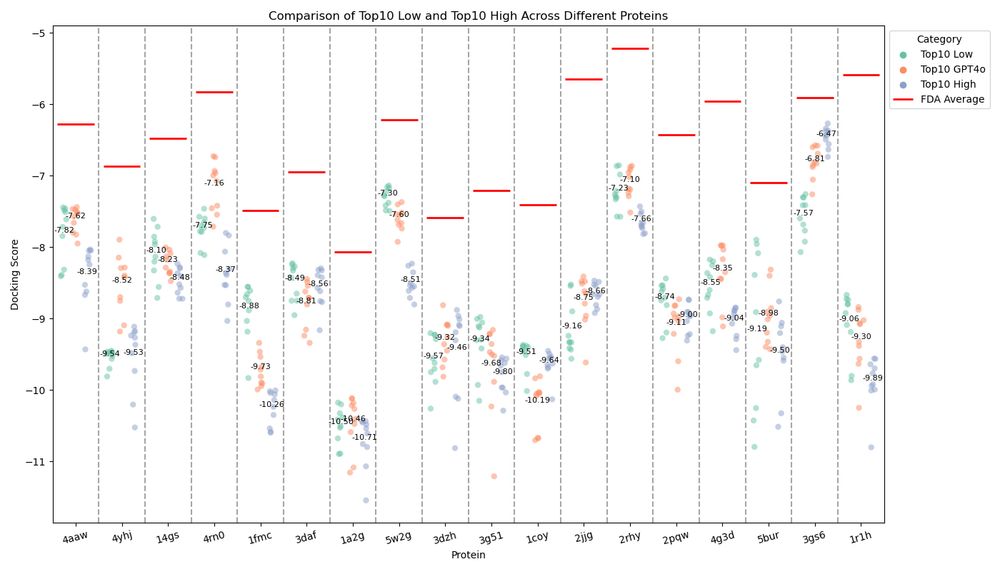
We wanted to generalize that for any new query and assess the effectiveness. 3/
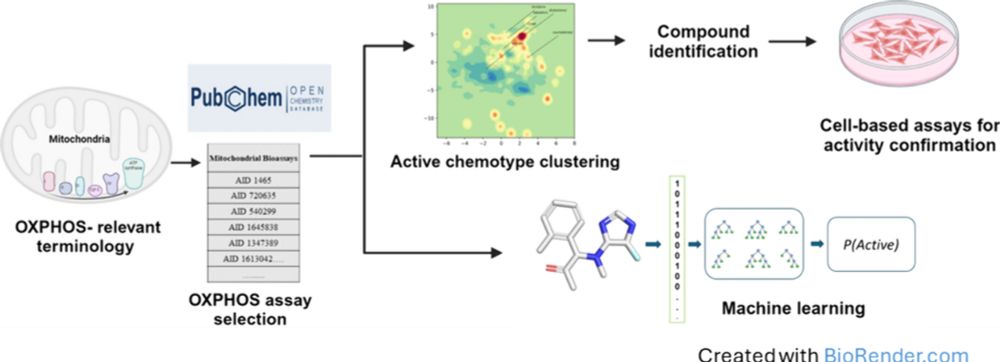
We wanted to generalize that for any new query and assess the effectiveness. 3/
There are now 1.7M PubChem BioAssays ranging in scale from a few tested molecules to high-throughput screens. 2/
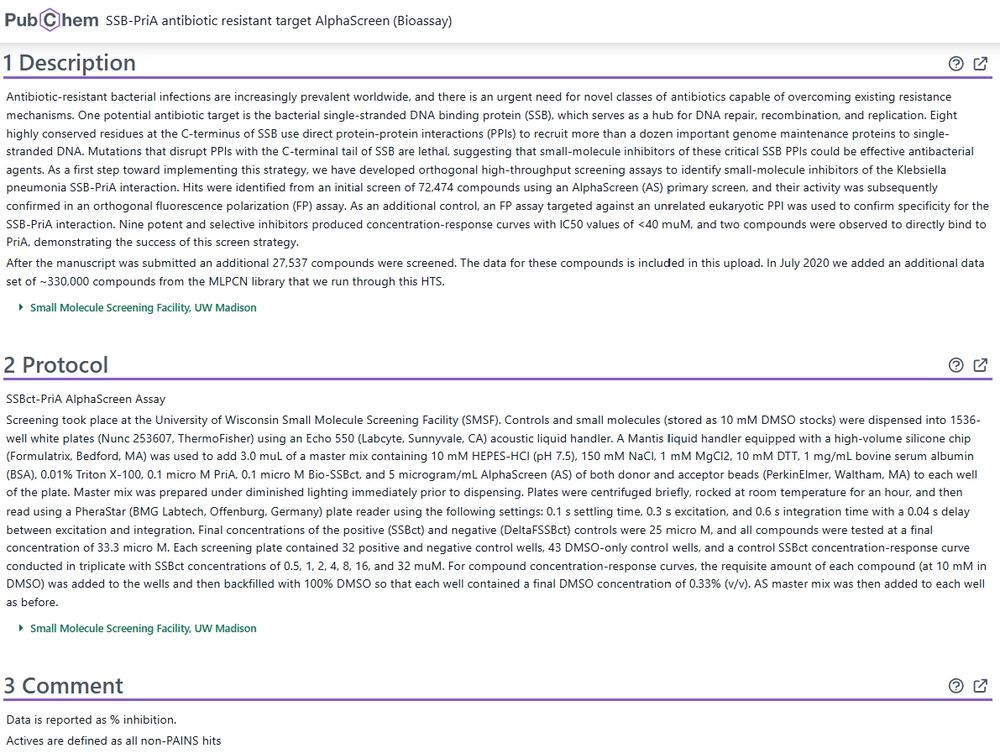
There are now 1.7M PubChem BioAssays ranging in scale from a few tested molecules to high-throughput screens. 2/

