MPAC uses biological pathway graphs to model DNA copy number and gene expression changes and infer activity states of all pathway members.

MPAC uses biological pathway graphs to model DNA copy number and gene expression changes and infer activity states of all pathway members.
Our collaboration with @anthonygitter.bsky.social is out in Nature Methods! We use synthetic data from molecular modeling to pretrain protein language models. Congrats to Sam Gelman and the team!
🔗 www.nature.com/articles/s41...

Our collaboration with @anthonygitter.bsky.social is out in Nature Methods! We use synthetic data from molecular modeling to pretrain protein language models. Congrats to Sam Gelman and the team!
🔗 www.nature.com/articles/s41...
(api.esmatlas.com/foldSequence... gives me Service Temporarily Unavailable)
(api.esmatlas.com/foldSequence... gives me Service Temporarily Unavailable)
doi.org/10.1038/s415...

doi.org/10.1038/s415...
ChemLML is a method for text-based conditional molecule generation that uses pretrained text models like SciBERT, Galactica, or T5.

ChemLML is a method for text-based conditional molecule generation that uses pretrained text models like SciBERT, Galactica, or T5.
Can protein language models help us fight viral outbreaks? Not yet. Here’s why 🧵👇
1/12

Can protein language models help us fight viral outbreaks? Not yet. Here’s why 🧵👇
1/12
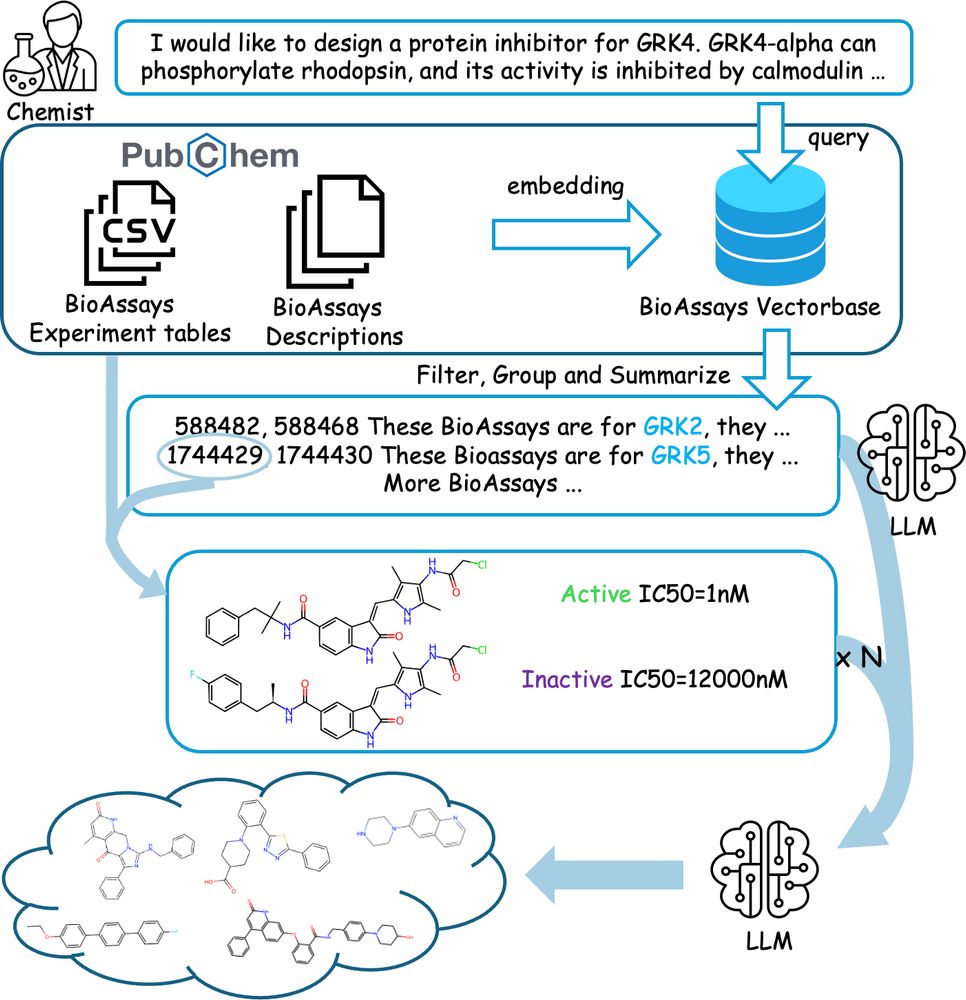

📄 academic.oup.com/nsr/article/...

📄 academic.oup.com/nsr/article/...
Explore these images and learn how protein research is changing our world. #NSFfunded #NSF75
pdb101.rcsb.org/lear...
Explore these images and learn how protein research is changing our world. #NSFfunded #NSF75
pdb101.rcsb.org/lear...

