MPAC uses biological pathway graphs to model DNA copy number and gene expression changes and infer activity states of all pathway members.

MPAC uses biological pathway graphs to model DNA copy number and gene expression changes and infer activity states of all pathway members.




There are now 1.7M PubChem BioAssays ranging in scale from a few tested molecules to high-throughput screens. 2/
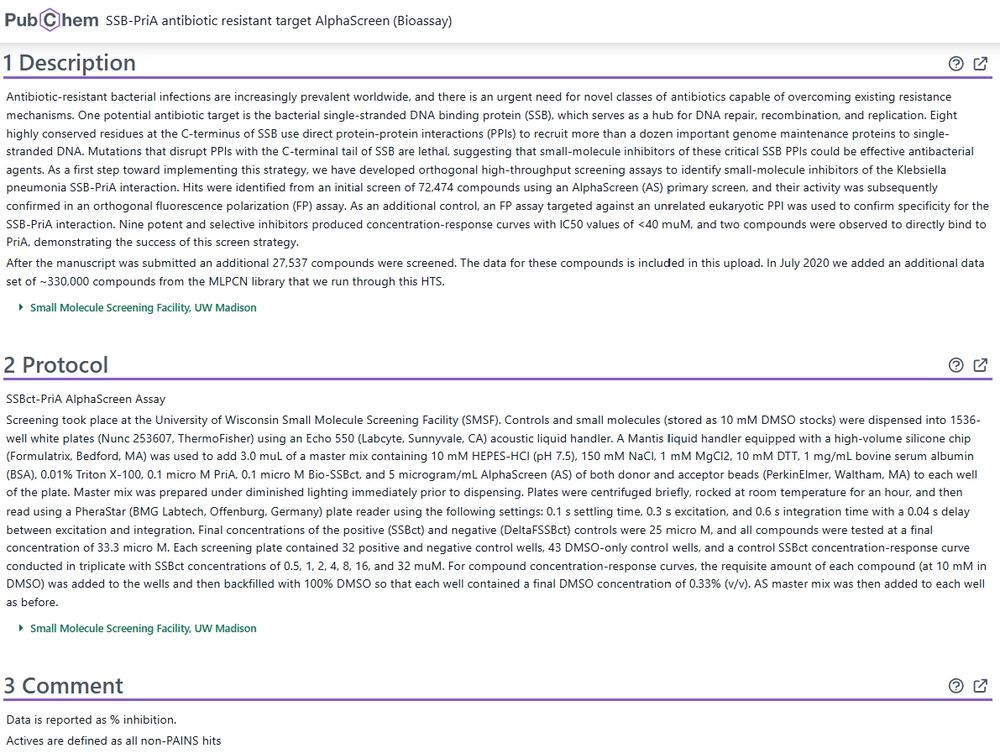
There are now 1.7M PubChem BioAssays ranging in scale from a few tested molecules to high-throughput screens. 2/