
🏥 64 cancer types
👥 2,372 participants
🧪 11,378 biospecimens
🧬 32 assays
📊 233,284 data files
Explore it now: data.humantumoratlas.org
🏥 64 cancer types
👥 2,372 participants
🧪 11,378 biospecimens
🧬 32 assays
📊 233,284 data files
Explore it now: data.humantumoratlas.org
Led by the excellent Ino de Bruijn and Milen Nikolov, it captures how we’ve made large-scale, multi-modal cancer data findable, explorable, & reusable.
www.nature.com/articles/s41...
Led by the excellent Ino de Bruijn and Milen Nikolov, it captures how we’ve made large-scale, multi-modal cancer data findable, explorable, & reusable.
www.nature.com/articles/s41...
lil.law.harvard.edu/blog/2025/02...
#openscience #datapreservation
lil.law.harvard.edu/blog/2025/02...
#openscience #datapreservation
www.nature.com/articles/s41...
www.nature.com/articles/s41...
—Integration of 5 omic modalities w/ spatial resolution at 20 -50 μm
—"a new era for characterizing tissue & cellular heterogeneity that single-modality studies alone could not reveal"
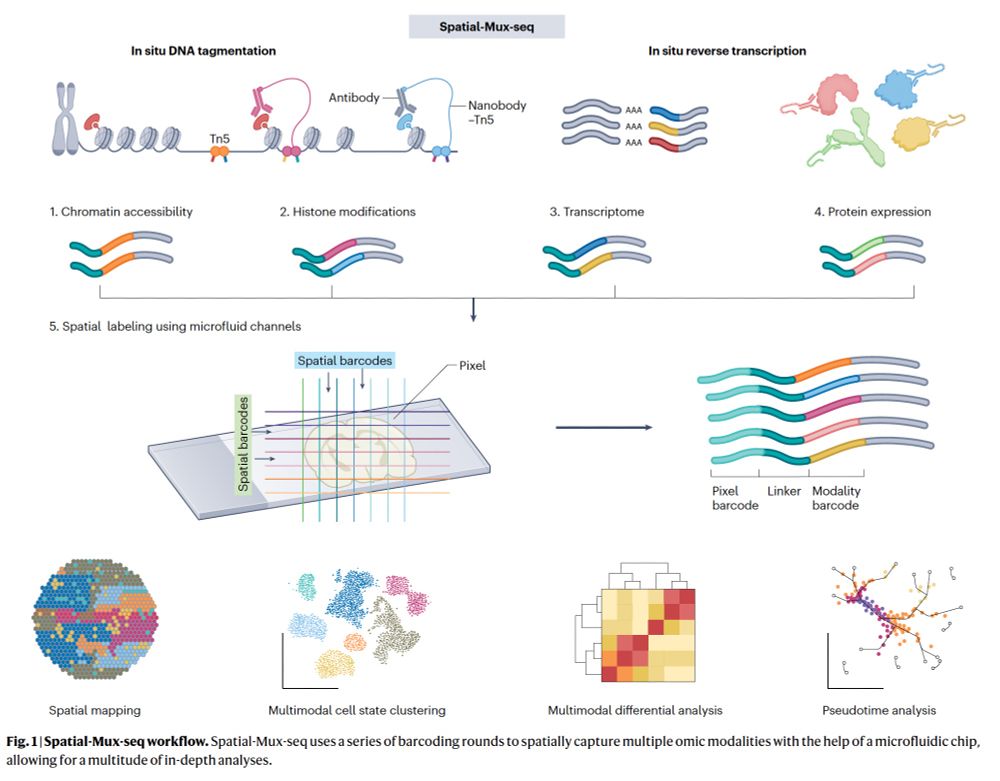
www.nature.com/articles/s41...
www.nature.com/articles/s41...
—Integration of 5 omic modalities w/ spatial resolution at 20 -50 μm
—"a new era for characterizing tissue & cellular heterogeneity that single-modality studies alone could not reveal"


