
👇 Read more

👇 Read more
pubs.rsc.org/en/content/a...
Fast, general-purpose metabolome analysis by mixed-mode liquid chromatography–mass spectrometry
Nicola Zamboni et al @nzamboni.bsky.social @alaaothman.bsky.social @ethz.ch
#MassSpec
pubs.rsc.org/en/content/a...
Fast, general-purpose metabolome analysis by mixed-mode liquid chromatography–mass spectrometry
Nicola Zamboni et al @nzamboni.bsky.social @alaaothman.bsky.social @ethz.ch
#MassSpec
Learn how Wikidata, which houses over 1.6 billion facts, advances education, innovation, and public institutions ➡️ wikimediafoundation.org/news/2025/10...

Learn how Wikidata, which houses over 1.6 billion facts, advances education, innovation, and public institutions ➡️ wikimediafoundation.org/news/2025/10...

Today, we are thrilled to announce that Wikidata has been recognized by the @DPGAlliance as a #DigitalPublicGood.

Today, we are thrilled to announce that Wikidata has been recognized by the @DPGAlliance as a #DigitalPublicGood.

Interested in improving our R tools for #MassSpectrometry data analysis and integrating them into Galaxy?
⏲️ 3 year position
📍 Bolzano, 🇮🇹
👉 apply if you like:
- #rstats SW development
- @bioconductor.bsky.social
- large-scale #metabolomics data analysis
- hiking ⛰️
🔗 bit.ly/46AMawx
Interested in improving our R tools for #MassSpectrometry data analysis and integrating them into Galaxy?
⏲️ 3 year position
📍 Bolzano, 🇮🇹
👉 apply if you like:
- #rstats SW development
- @bioconductor.bsky.social
- large-scale #metabolomics data analysis
- hiking ⛰️
🔗 bit.ly/46AMawx
And it supports ORCID to link to authors, "title in HTML", mul, and […]
And it supports ORCID to link to authors, "title in HTML", mul, and […]
It places the scalability of MS-based metabolomics for natural extracts as a central challenge rather than a side issue.
#metabolomics #CompMetabolomics #naturalproducts #discovery #libraries
pubs.rsc.org/en/content/a...

It places the scalability of MS-based metabolomics for natural extracts as a central challenge rather than a side issue.
In this work, we identified small molecule effectors modulating the activity of transcription factors in E. coli.
Using metabolomics + transcriptomics, we could parse the entire regulatory network and predict signal molecules for 41 TFs!
www.embopress.org/doi/full/10....

In this work, we identified small molecule effectors modulating the activity of transcription factors in E. coli.
Using metabolomics + transcriptomics, we could parse the entire regulatory network and predict signal molecules for 41 TFs!
www.embopress.org/doi/full/10....
doi.org/10.26434/che...

doi.org/10.26434/che...

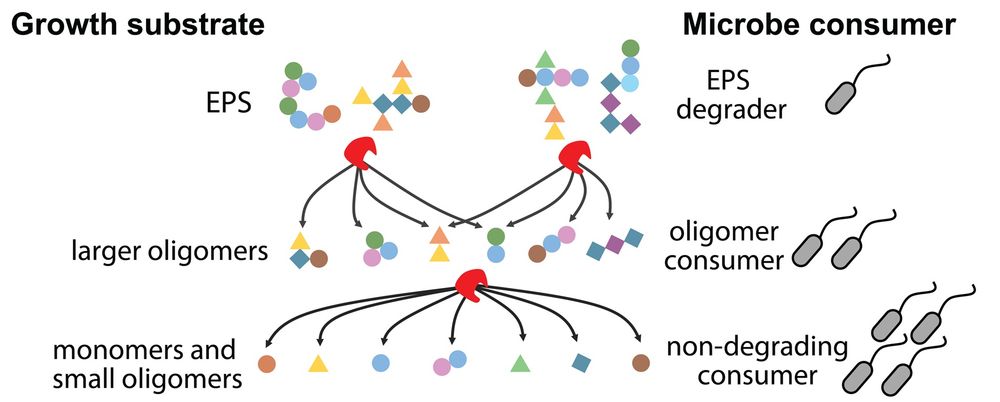
Looking forward to it, and particularly talking about the InChI for inorganics and trying that in @wikidata :) See https://doi.org/10.26434/chemrxiv-2025-53n0w
And also the nano InChI, see […]
Looking forward to it, and particularly talking about the InChI for inorganics and trying that in @wikidata :) See https://doi.org/10.26434/chemrxiv-2025-53n0w
And also the nano InChI, see […]
www.nature.com/articles/s41...
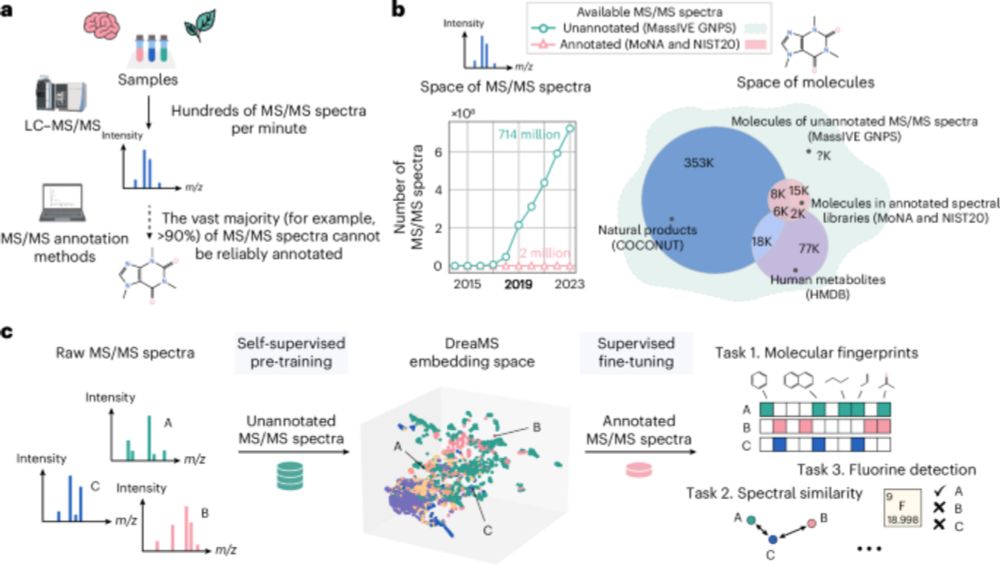
www.nature.com/articles/s41...
pontrellilab.sites.vib.be/en

pontrellilab.sites.vib.be/en
lnkd.in/erh_eeTy
lnkd.in/erh_eeTy
lnkd.in/ea8gzBzG
lnkd.in/ea8gzBzG
lnkd.in/eZZrkuFb
lnkd.in/eZZrkuFb

You'll work at WUR in the Netherlands, ranked #3 in environ. sciences, #1 in agricultural science, #38 in life sciences (QS).
Apply here:
www.wur.nl/nl/vacature/...

You'll work at WUR in the Netherlands, ranked #3 in environ. sciences, #1 in agricultural science, #38 in life sciences (QS).
Apply here:
www.wur.nl/nl/vacature/...
Dr. Robinson and her team at @eawag.bsky.social focus on Microbial Specialized Metabolism, exploring how microbes 🦠 and their enzymes help degrade pollutants. Her research is crucial for cleaning up contaminants & improving food system sustainability.

Dr. Robinson and her team at @eawag.bsky.social focus on Microbial Specialized Metabolism, exploring how microbes 🦠 and their enzymes help degrade pollutants. Her research is crucial for cleaning up contaminants & improving food system sustainability.

