Begin your academic career with the expert scientists, technology, and facilities at the Earlham Institute in Norwich.
buff.ly/327FNrX
#genomics #findaphd #AcademicSky
Begin your academic career with the expert scientists, technology, and facilities at the Earlham Institute in Norwich.
buff.ly/327FNrX
#genomics #findaphd #AcademicSky
🔗 buff.ly/sUekTX5

🔗 buff.ly/sUekTX5


📃 Check out the preprint here: www.biorxiv.org/content/10.1...


📃 Check out the preprint here: www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
🖥️🧬
@whatchamacaulay.bsky.social




🖥️🧬
@whatchamacaulay.bsky.social
Thanks @mattbawn.bsky.social for organising 👍
Me, Matt, Beth & Aleyda had a great time 😎

Thanks @mattbawn.bsky.social for organising 👍
Me, Matt, Beth & Aleyda had a great time 😎
Details and programme below 👇
buff.ly/yUNXYK1
🖥️ 🧬
#scRNAseq #Bioinformatics #Workshop

Details and programme below 👇
buff.ly/yUNXYK1
🖥️ 🧬
#scRNAseq #Bioinformatics #Workshop
At the Earlham Institute, we use the latest #singlecell and #spatial methods to understand variation of #geneexpression at the cellular level.
🔗 buff.ly/twqX8ie
@whatchamacaulay.bsky.social



At the Earlham Institute, we use the latest #singlecell and #spatial methods to understand variation of #geneexpression at the cellular level.
🔗 buff.ly/twqX8ie
@whatchamacaulay.bsky.social
🦠 Environmental microbes
🧠 Healthy human development and ageing
🌱 Plant development
🧬 Human pathology
↔️ Cell dynamics
🖥️ 🧬 🧪
🦠 Environmental microbes
🧠 Healthy human development and ageing
🌱 Plant development
🧬 Human pathology
↔️ Cell dynamics
🖥️ 🧬 🧪
See you all in July (9th - 11th)
🌞🧬🧬👍 #Genomics
- Early bird registration closure 16th May
- Abstract submission closure 23rd May
- Registration closure 26th June
www.genomescience.org.uk

See you all in July (9th - 11th)
🌞🧬🧬👍 #Genomics
- Early bird registration closure 16th May
- Abstract submission closure 23rd May
- Registration closure 26th June
www.genomescience.org.uk
Join us @earlhaminst.bsky.social on the 18th and 19th of June to discuss all aspects of #singlecell and #spatial #genomics - from new technologies to applications in microbiology, plants, animals and human!
More info and registration here: shorturl.at/2R9ek
Join us @earlhaminst.bsky.social on the 18th and 19th of June to discuss all aspects of #singlecell and #spatial #genomics - from new technologies to applications in microbiology, plants, animals and human!
More info and registration here: shorturl.at/2R9ek
Join us @earlhaminst.bsky.social on the 18th and 19th of June to discuss all aspects of #singlecell and #spatial #genomics - from new technologies to applications in microbiology, plants, animals and human!
More info and registration here: shorturl.at/2R9ek

Join us @earlhaminst.bsky.social on the 18th and 19th of June to discuss all aspects of #singlecell and #spatial #genomics - from new technologies to applications in microbiology, plants, animals and human!
More info and registration here: shorturl.at/2R9ek
www.genomescience.org.uk
There's a great programme developing - covering Microbial / Plant / Animal / Human #Genomics with a major focus on emerging and transformative technologies!
www.genomescience.org.uk
There's a great programme developing - covering Microbial / Plant / Animal / Human #Genomics with a major focus on emerging and transformative technologies!
Read more on the work, first published in ISME Comms: jgi.doe.gov/user-science...
@berkeleylab.lbl.gov @danudwary.secondarymetabolism.com @axelvisel.bsky.social

Read more on the work, first published in ISME Comms: jgi.doe.gov/user-science...
@berkeleylab.lbl.gov @danudwary.secondarymetabolism.com @axelvisel.bsky.social

sanger.wd103.myworkdayjobs.com/WellcomeSang...
Incredibly exciting datasets on host-microbe interactions in the lung.
Pls RT and reach out if you are interested.
www.genomescience.org.uk
There's a great programme developing - covering Microbial / Plant / Animal / Human #Genomics with a major focus on emerging and transformative technologies!
www.genomescience.org.uk
There's a great programme developing - covering Microbial / Plant / Animal / Human #Genomics with a major focus on emerging and transformative technologies!
Submissions for small talks are open until April 30, 2025.
All other applications for this conference are due July 13, 2025.
To Apply: bit.ly/ApplyforGRC

Submissions for small talks are open until April 30, 2025.
All other applications for this conference are due July 13, 2025.
To Apply: bit.ly/ApplyforGRC
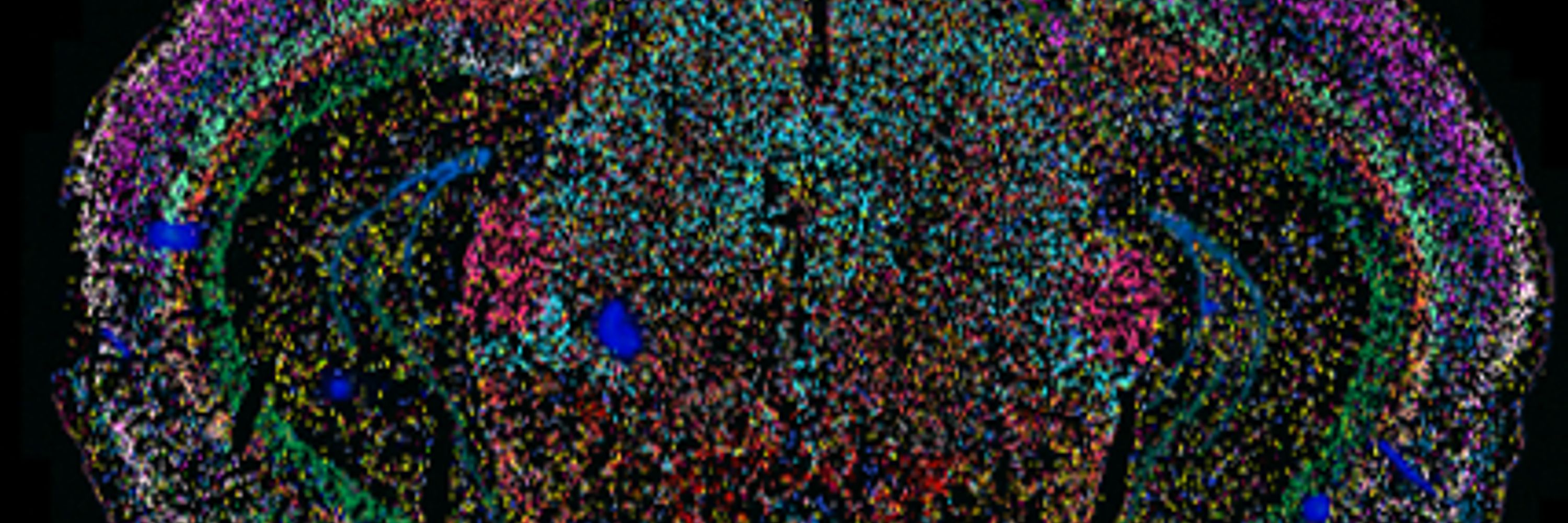
@deboradesantis.bsky.social recently joined @whatchamacaulay.bsky.social Group to further develop her skills in single-nuclei RNA sequencing. 👇

@deboradesantis.bsky.social recently joined @whatchamacaulay.bsky.social Group to further develop her skills in single-nuclei RNA sequencing. 👇
theconversation.com/ignored-blam...

theconversation.com/ignored-blam...

