We present "Transferable Generative Models Bridge Femtosecond to Nanosecond Time-Step Molecular Dynamics,"

We present "Transferable Generative Models Bridge Femtosecond to Nanosecond Time-Step Molecular Dynamics,"
doi.org/10.1016/j.ai...
Work led by Sofia Larsson and Miranda Carlsson, with @rbeckmann.bsky.social and Filip Miljković (AZ)!
#compchem #chemsky

doi.org/10.1016/j.ai...
Work led by Sofia Larsson and Miranda Carlsson, with @rbeckmann.bsky.social and Filip Miljković (AZ)!
#compchem #chemsky
@janstuehmer.bsky.social, @arnauddoucet.bsky.social, @marcocuturi.bsky.social , Marta Betcke,
Elena Agliari, Beatriz Seoane, Alessandro Ingrosso

@janstuehmer.bsky.social, @arnauddoucet.bsky.social, @marcocuturi.bsky.social , Marta Betcke,
Elena Agliari, Beatriz Seoane, Alessandro Ingrosso
@janstuehmer.bsky.social, @arnauddoucet.bsky.social, @marcocuturi.bsky.social , Marta Betcke,
Elena Agliari, Beatriz Seoane, Alessandro Ingrosso

See you tomorrow at poster 13, 10-12:30.
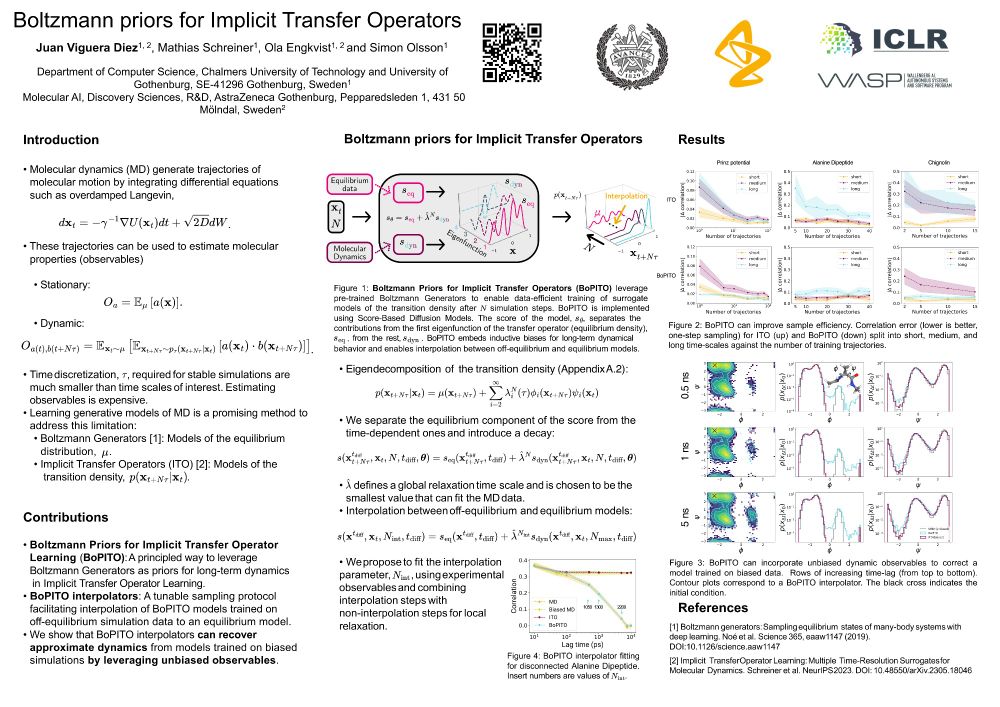
See you tomorrow at poster 13, 10-12:30.
Protein assembly in membranes is crucial yet elusive. Why steer when you can just observe? We introduce a bias-free simulation method that captures the full picture of transmembrane dimerization—free energies, mechanisms, and rates!
pubs.acs.org/doi/10.1021/...
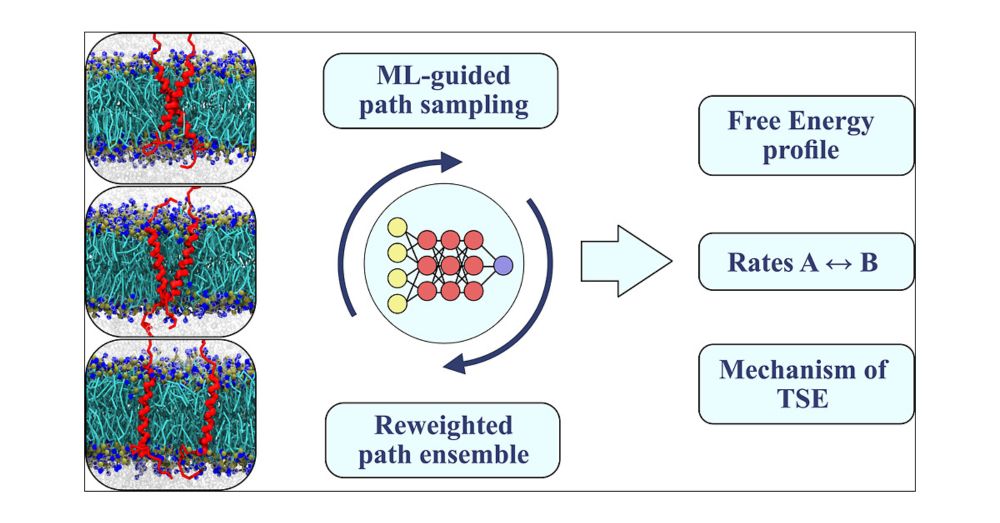
Protein assembly in membranes is crucial yet elusive. Why steer when you can just observe? We introduce a bias-free simulation method that captures the full picture of transmembrane dimerization—free energies, mechanisms, and rates!
pubs.acs.org/doi/10.1021/...
Come join us on an exciting joint PhD project on ML for protein function with @kailalab.bsky.social -- We are looking for a strong quantitative candidate with prior experience in ML. The position is based in Gothenburg and is fully funded for 5 years.
www.chalmers.se/en/about-cha...
Come join us on an exciting joint PhD project on ML for protein function with @kailalab.bsky.social -- We are looking for a strong quantitative candidate with prior experience in ML. The position is based in Gothenburg and is fully funded for 5 years.
www.chalmers.se/en/about-cha...
A novel machine-learning-based force field for protein thermodynamics. It uses an all-heavy-atom approach without hydrogens to simplify simulations while maintaining key dynamics.
pubs.acs.org/doi/10.1021/...

A novel machine-learning-based force field for protein thermodynamics. It uses an all-heavy-atom approach without hydrogens to simplify simulations while maintaining key dynamics.
pubs.acs.org/doi/10.1021/...