If we can get vendor buy-in to an open, common format, it could bring more computational folks into the field. How can these folks be excited about #proteomics when its hard to even read the already complicated data?
pubs.acs.org/doi/10.1021/...

If we can get vendor buy-in to an open, common format, it could bring more computational folks into the field. How can these folks be excited about #proteomics when its hard to even read the already complicated data?
pubs.acs.org/doi/10.1021/...



Read the Editorial by EiC Daniel Gorelick @danielgorelick.bsky.social at: bit.ly/4kYD1mL

collab with @kusterlab.bsky.social and @bendavisgroup.bsky.social
www.mcponline.org/article/S153...
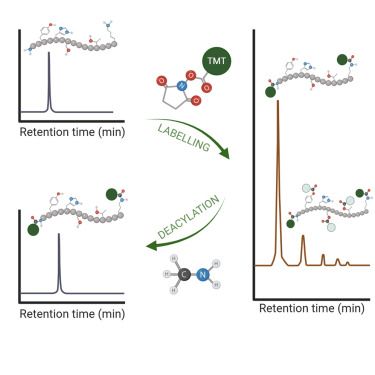
collab with @kusterlab.bsky.social and @bendavisgroup.bsky.social
www.mcponline.org/article/S153...
some processing is usually necessary, overcooking makes it bland, highly over-processed foods are unhealthy, and the quality of the ingredients matters.
some processing is usually necessary, overcooking makes it bland, highly over-processed foods are unhealthy, and the quality of the ingredients matters.
#TheraNova, #ENABLE, #Proxidrugs