
, data science and philosophy of science.
* Post-doc (Structural Bioinformatics) / Mendel University, Brno - Czech Republic.
* Research interest: Proteins side chains and Rotamers.

The proteomic code: Novel amino acid residue pairing models “encode” protein folding and protein-protein interactions:
www.sciencedirect.com/science/arti...
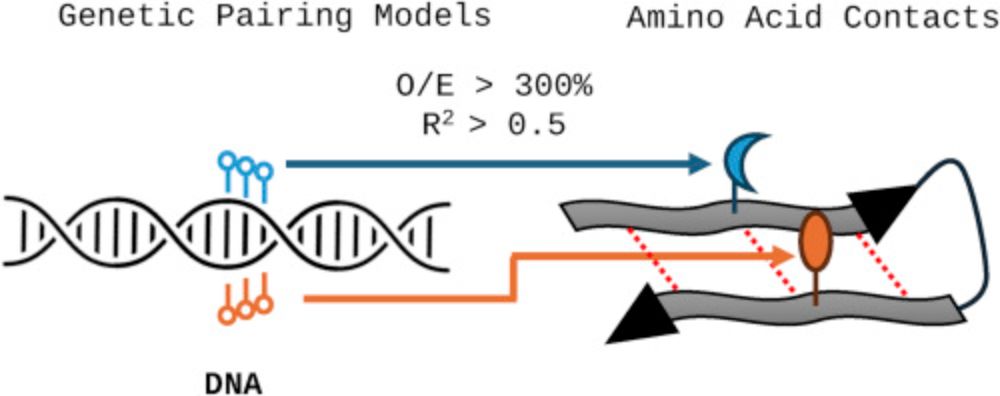
The proteomic code: Novel amino acid residue pairing models “encode” protein folding and protein-protein interactions:
www.sciencedirect.com/science/arti...

