go.nature.com/4iX9R6N
Free to read here: rdcu.be/e1t9M
#worldntdday
go.nature.com/4iX9R6N
Free to read here: rdcu.be/e1t9M
#worldntdday
Read it here! www.biorxiv.org/content/10.6...

Read it here! www.biorxiv.org/content/10.6...


go.nature.com/4iX9R6N

go.nature.com/4iX9R6N

🔗 doi.org/10.1093/molbev/msaf253
#evobio #molbio #phylogenetics

🔗 doi.org/10.1093/molbev/msaf253
#evobio #molbio #phylogenetics
www.eurosurveillance.org/content/10.2...
Wonderful collaborative effort conducted in the context of mood-h2020.eu
Read the thread below 👇
www.eurosurveillance.org/content/10.2...
Wonderful collaborative effort conducted in the context of mood-h2020.eu
Read the thread below 👇
Only 1/3 of contacts shared the same strain 🤯, so most people misperceived where they got infected.
@thibaut-jonathan.bsky.social and team show how to assess contact tracing accuracy in the future!
#IDSky #EpiSky

Only 1/3 of contacts shared the same strain 🤯, so most people misperceived where they got infected.
@thibaut-jonathan.bsky.social and team show how to assess contact tracing accuracy in the future!
#IDSky #EpiSky

Details and instructions for installing on the BEAST website: beast.community

Details and instructions for installing on the BEAST website: beast.community
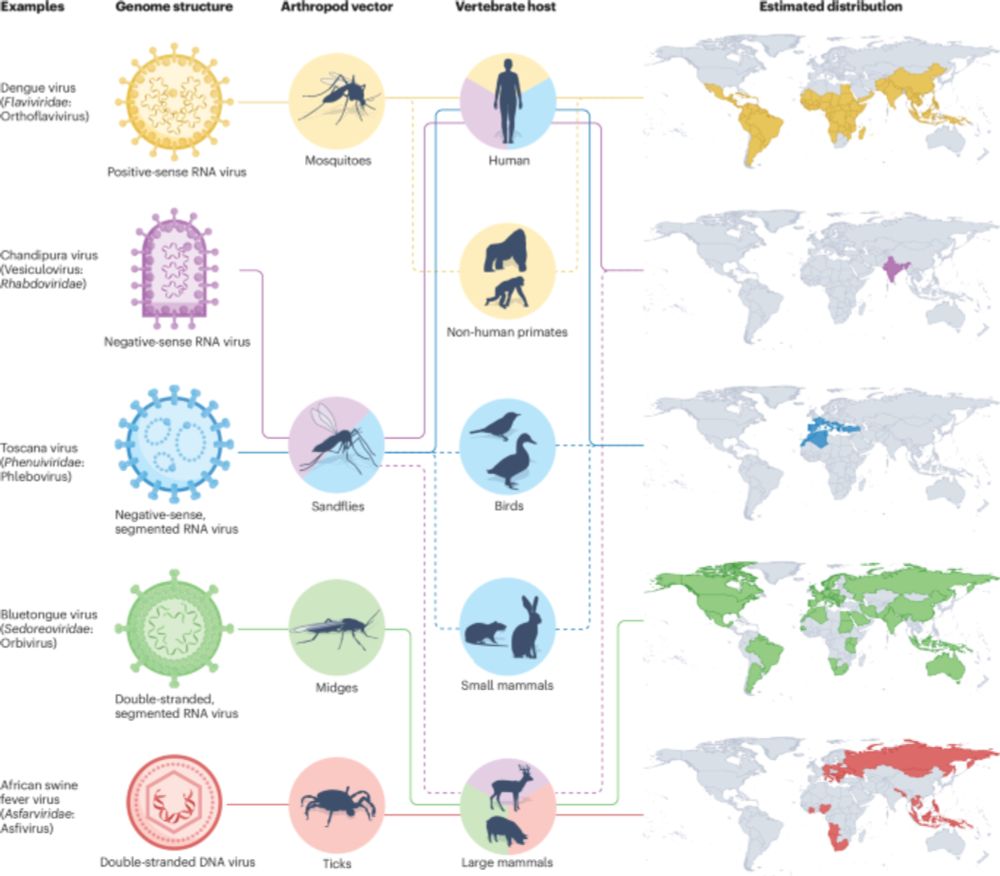
www.nature.com/articles/s41...
Great work by Verity Hill and team
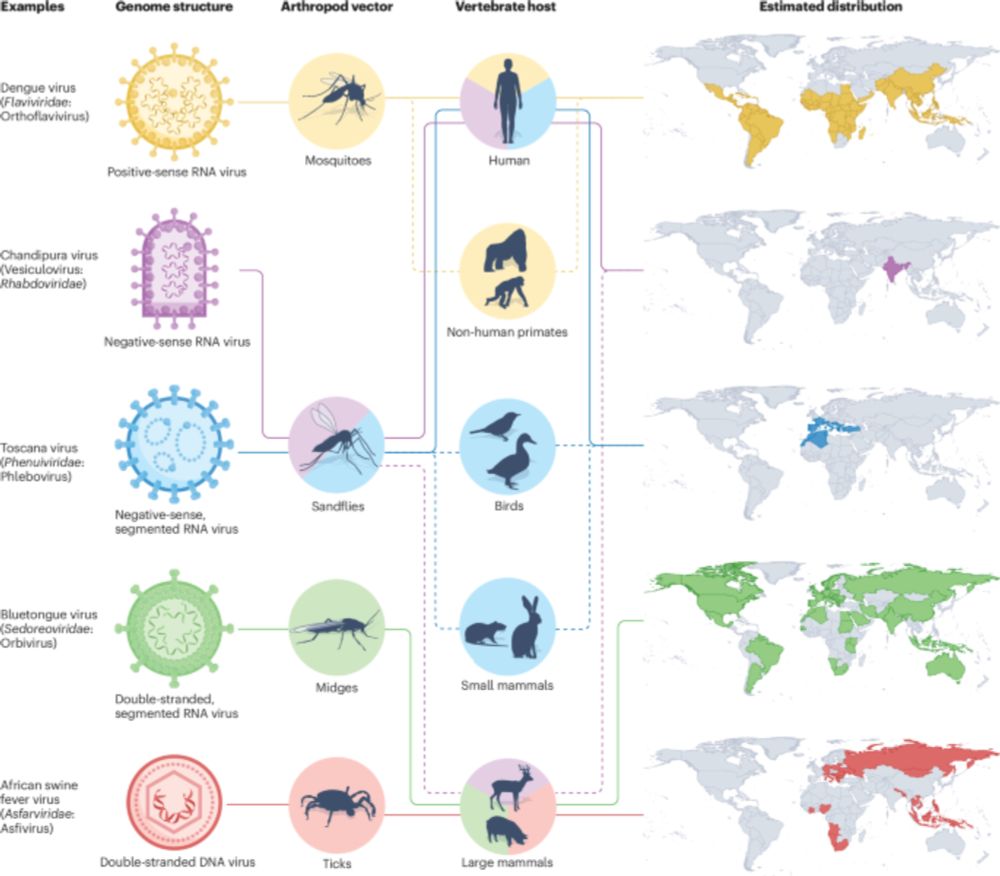
www.nature.com/articles/s41...
Great work by Verity Hill and team
📖 👉 rdcu.be/epH2U
Short 🧵

📖 👉 rdcu.be/epH2U
Short 🧵
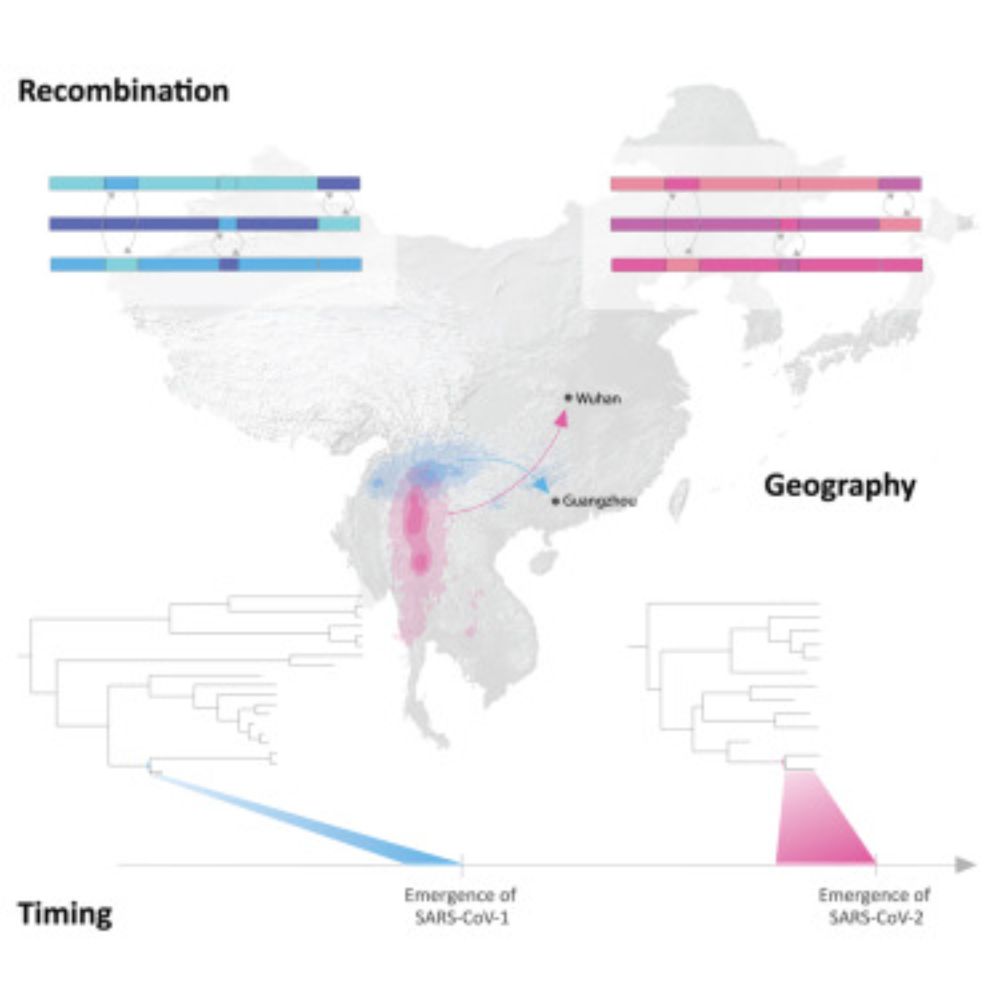
www.cell.com/cell/fulltex...
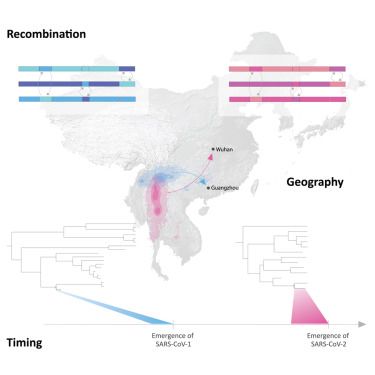
www.cell.com/cell/fulltex...
👉 bit.ly/4m3Fb4Y
@mariusgilbert.bsky.social @sdellicour.bsky.social

👉 bit.ly/4m3Fb4Y
@mariusgilbert.bsky.social @sdellicour.bsky.social

Our latest study on HPAI in France and risk mapping has just been published doi.org/10.1371/jour...
@epidesa.bsky.social @inrae-france.bsky.social @anses-fr.bsky.social ENVT ULB
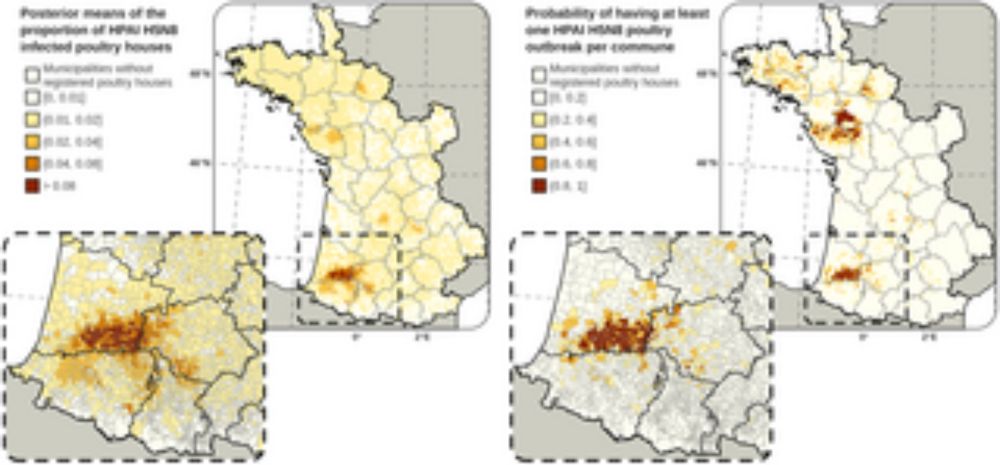
Our latest study on HPAI in France and risk mapping has just been published doi.org/10.1371/jour...
@epidesa.bsky.social @inrae-france.bsky.social @anses-fr.bsky.social ENVT ULB

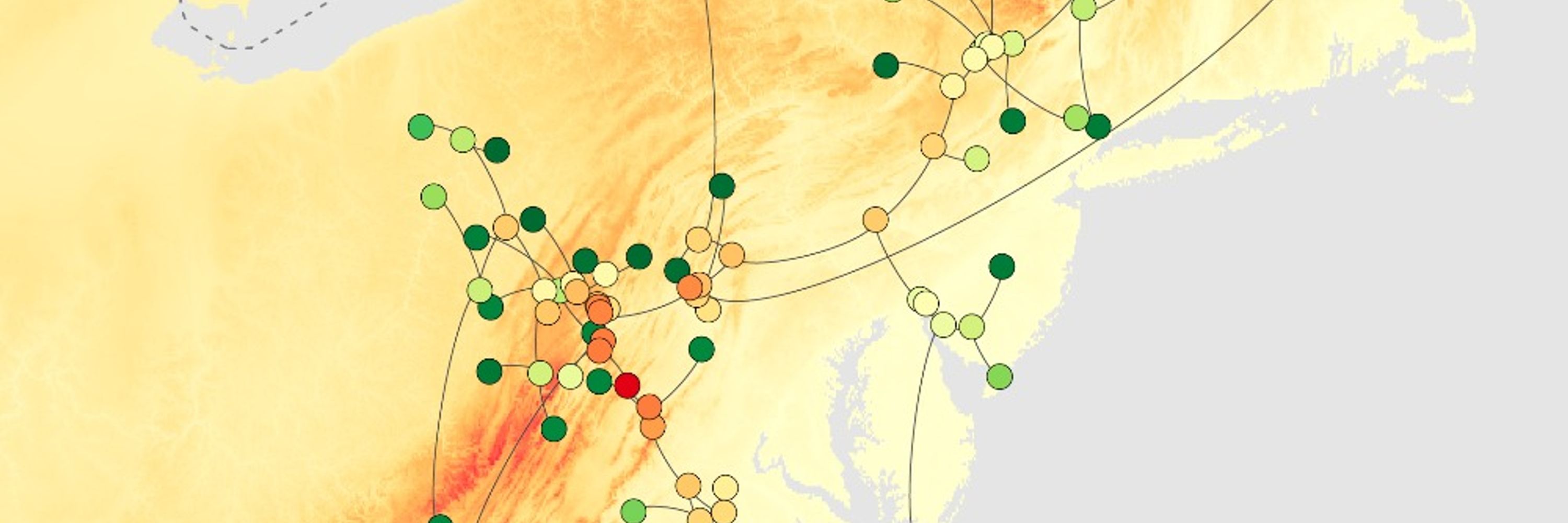
Modeling the velocity of evolving lineages and predicting dispersal patterns
www.pnas.org/doi/10.1073/...
Modeling the velocity of evolving lineages and predicting dispersal patterns
www.pnas.org/doi/10.1073/...
www.statnews.com/2024/12/02/b...

www.statnews.com/2024/12/02/b...
More and more human cases are occurring, along with mutations that indicate further human adaptation.
We need to treat these cases like a warning, because that is what they are.
www.theguardian.com/world/2024/n...
More and more human cases are occurring, along with mutations that indicate further human adaptation.
We need to treat these cases like a warning, because that is what they are.
www.theguardian.com/world/2024/n...


