Sam Lipworth
@samlipworth.bsky.social
Academic Clinical Fellow and ID/Micro registrar in Oxford
🔔🔔Post-doc in microbial genomics with @modmedmicro.bsky.social🧬 - deadline tomorrow! Big genomic and clinical datasets to play with, friendly and very multidisciplinary team!
my.corehr.com/pls/uoxrecru...
my.corehr.com/pls/uoxrecru...
Job Details
my.corehr.com
October 15, 2025 at 10:58 AM
🔔🔔Post-doc in microbial genomics with @modmedmicro.bsky.social🧬 - deadline tomorrow! Big genomic and clinical datasets to play with, friendly and very multidisciplinary team!
my.corehr.com/pls/uoxrecru...
my.corehr.com/pls/uoxrecru...
Reposted by Sam Lipworth
Our recent paper on rifampicin resistant subpopulations in M. tuberculosis (M. tb) has been published at JAC-antimicrobial resistance.
I am really happy to see this work published just hours before submitting my DPhil thesis! 🔗👇
doi.org/10.1093/jaca...
I am really happy to see this work published just hours before submitting my DPhil thesis! 🔗👇
doi.org/10.1093/jaca...

Subpopulations in clinical samples of M. tuberculosis can give rise to rifampicin resistance and shed light on how resistance is acquired
AbstractObjectives. WGS has become a key tool for diagnosing Mycobacterium tuberculosis infections, but discrepancies between genotypic and phenotypic drug
doi.org
October 13, 2025 at 4:40 PM
Our recent paper on rifampicin resistant subpopulations in M. tuberculosis (M. tb) has been published at JAC-antimicrobial resistance.
I am really happy to see this work published just hours before submitting my DPhil thesis! 🔗👇
doi.org/10.1093/jaca...
I am really happy to see this work published just hours before submitting my DPhil thesis! 🔗👇
doi.org/10.1093/jaca...
We have a fully funded PhD studentship available @modmedmicro.bsky.social in Oxford to work on a really cool project looking at population heterogeneity in bloodstream infections www.ndm.ox.ac.uk/study/dphil-... Please spread the word/get in touch if you'd like to chat.
DPhil project themes
www.ndm.ox.ac.uk
October 8, 2025 at 7:11 PM
We have a fully funded PhD studentship available @modmedmicro.bsky.social in Oxford to work on a really cool project looking at population heterogeneity in bloodstream infections www.ndm.ox.ac.uk/study/dphil-... Please spread the word/get in touch if you'd like to chat.
Reposted by Sam Lipworth
New research article
Estimating the association of antimicrobial resistance genes with minimum inhibitory concentration in Escherichia coli: an observational study
www.thelancet.com/journals/lan...
#IDSky #ClinMicro #AMR #WGS #OpenAccess #OA
Estimating the association of antimicrobial resistance genes with minimum inhibitory concentration in Escherichia coli: an observational study
www.thelancet.com/journals/lan...
#IDSky #ClinMicro #AMR #WGS #OpenAccess #OA

October 8, 2025 at 8:28 AM
New research article
Estimating the association of antimicrobial resistance genes with minimum inhibitory concentration in Escherichia coli: an observational study
www.thelancet.com/journals/lan...
#IDSky #ClinMicro #AMR #WGS #OpenAccess #OA
Estimating the association of antimicrobial resistance genes with minimum inhibitory concentration in Escherichia coli: an observational study
www.thelancet.com/journals/lan...
#IDSky #ClinMicro #AMR #WGS #OpenAccess #OA
Pleased to see this out today in @lancetmicrobe.bsky.social www.sciencedirect.com/science/arti.... We estimate the independent effects of AMR genes on MIC at a drug-bug level and challenge the idea that resistance is a binary phenomenon.

Estimating the association of antimicrobial resistance genes with minimum inhibitory concentration in Escherichia coli: an observational study
Surveillance and prediction of antibiotic resistance in Escherichia coli relies on curated databases of genes and mutations. We aimed to quantify the …
www.sciencedirect.com
October 8, 2025 at 6:25 AM
Pleased to see this out today in @lancetmicrobe.bsky.social www.sciencedirect.com/science/arti.... We estimate the independent effects of AMR genes on MIC at a drug-bug level and challenge the idea that resistance is a binary phenomenon.
Reposted by Sam Lipworth
From Pipettes to Pipelines: Greening a Multidisciplinary Research Lab.
@dotnagy.bsky.social writes about the experiences from the Modernising Medical Microbiology (MMM) research group on engaging wet lab, computer lab, and office teams on the #sustainability agenda.
#GreenLab #SustainableScience
@dotnagy.bsky.social writes about the experiences from the Modernising Medical Microbiology (MMM) research group on engaging wet lab, computer lab, and office teams on the #sustainability agenda.
#GreenLab #SustainableScience

From Pipettes to Pipelines — Oxford Climate Society
Dorottya Nagy writes about the experiences from the Modernising Medical Microbiology (MMM) Unit research group on engaging wet lab, computer lab, and office teams on the sustainability agenda.
oxfordclimatesociety.com
September 30, 2025 at 4:45 PM
From Pipettes to Pipelines: Greening a Multidisciplinary Research Lab.
@dotnagy.bsky.social writes about the experiences from the Modernising Medical Microbiology (MMM) research group on engaging wet lab, computer lab, and office teams on the #sustainability agenda.
#GreenLab #SustainableScience
@dotnagy.bsky.social writes about the experiences from the Modernising Medical Microbiology (MMM) research group on engaging wet lab, computer lab, and office teams on the #sustainability agenda.
#GreenLab #SustainableScience
Reposted by Sam Lipworth
Now published in @natcomms.nature.com 🎉
www.nature.com/articles/s41...
With Gillian Rodger, @nstoesser.bsky.social, @samlipworth.bsky.social, @stat-sarah.bsky.social, and many others!
www.nature.com/articles/s41...
With Gillian Rodger, @nstoesser.bsky.social, @samlipworth.bsky.social, @stat-sarah.bsky.social, and many others!
September 30, 2025 at 4:21 PM
Now published in @natcomms.nature.com 🎉
www.nature.com/articles/s41...
With Gillian Rodger, @nstoesser.bsky.social, @samlipworth.bsky.social, @stat-sarah.bsky.social, and many others!
www.nature.com/articles/s41...
With Gillian Rodger, @nstoesser.bsky.social, @samlipworth.bsky.social, @stat-sarah.bsky.social, and many others!
Reposted by Sam Lipworth
PhD opportunity to come and work in Glasgow, developing advanced tissue culture models to study Staphylococcus aureus colonisation and infection #microsky #PhD #DTPprogram
Enquiries from potential applicants are welcome.
www.gla.ac.uk/postgraduate...
www.findaphd.com/phds/program...
Enquiries from potential applicants are welcome.
www.gla.ac.uk/postgraduate...
www.findaphd.com/phds/program...
University of Glasgow - Postgraduate study - Centres for Doctoral Training - NorthWest Biosciences - Our Projects - Understanding Pathogens, from Molecules to Phenotypes - Justine Rudkin
www.gla.ac.uk
September 30, 2025 at 12:52 PM
PhD opportunity to come and work in Glasgow, developing advanced tissue culture models to study Staphylococcus aureus colonisation and infection #microsky #PhD #DTPprogram
Enquiries from potential applicants are welcome.
www.gla.ac.uk/postgraduate...
www.findaphd.com/phds/program...
Enquiries from potential applicants are welcome.
www.gla.ac.uk/postgraduate...
www.findaphd.com/phds/program...
Reposted by Sam Lipworth
Now published, our tool to run (almost) all biological models interactively in your web browser
Paper: academic.oup.com/bioinformati...
Website: biomodels.bacpop.org
Code: github.com/bacpop/SBMLt...
Paper: academic.oup.com/bioinformati...
Website: biomodels.bacpop.org
Code: github.com/bacpop/SBMLt...
September 29, 2025 at 9:49 AM
Now published, our tool to run (almost) all biological models interactively in your web browser
Paper: academic.oup.com/bioinformati...
Website: biomodels.bacpop.org
Code: github.com/bacpop/SBMLt...
Paper: academic.oup.com/bioinformati...
Website: biomodels.bacpop.org
Code: github.com/bacpop/SBMLt...
Reposted by Sam Lipworth
To summarise our recent pre-print: Autocycler, the automated consensus assembler, when used with Nanopore long-read only Enterobacterales assemblies, produces more complete chromosomes and plasmids, with an accuracy comparable to hybrid assemblies.

September 29, 2025 at 6:57 AM
To summarise our recent pre-print: Autocycler, the automated consensus assembler, when used with Nanopore long-read only Enterobacterales assemblies, produces more complete chromosomes and plasmids, with an accuracy comparable to hybrid assemblies.
Reposted by Sam Lipworth
Delighted to see our paper studying the evolution of plasmids over the last 100 years, now out! Years of work by Adrian Cazares, also Nick Thomson @sangerinstitute.bsky.social - this version much improved over the preprint. Final version should be open access, apols.
Thread 1/n
Thread 1/n

September 25, 2025 at 9:29 PM
Delighted to see our paper studying the evolution of plasmids over the last 100 years, now out! Years of work by Adrian Cazares, also Nick Thomson @sangerinstitute.bsky.social - this version much improved over the preprint. Final version should be open access, apols.
Thread 1/n
Thread 1/n
Reposted by Sam Lipworth
Pleased to see this pre-printed, highlighting the completeness/accuracy of @nanoporetech.com long-read genome assembly for clinical Enterobacterales: www.biorxiv.org/content/10.1...
Thanks to colleagues @modmedmicro.bsky.social, @ukhsa.bsky.social, @genewiz.bsky.social and @oxfordbrc.bsky.social!
Thanks to colleagues @modmedmicro.bsky.social, @ukhsa.bsky.social, @genewiz.bsky.social and @oxfordbrc.bsky.social!
September 25, 2025 at 8:48 AM
Pleased to see this pre-printed, highlighting the completeness/accuracy of @nanoporetech.com long-read genome assembly for clinical Enterobacterales: www.biorxiv.org/content/10.1...
Thanks to colleagues @modmedmicro.bsky.social, @ukhsa.bsky.social, @genewiz.bsky.social and @oxfordbrc.bsky.social!
Thanks to colleagues @modmedmicro.bsky.social, @ukhsa.bsky.social, @genewiz.bsky.social and @oxfordbrc.bsky.social!
Reposted by Sam Lipworth
Ever run `install.packages()` and wish it were faster, smarter, and more reliable?
The {pak} package speeds things up with parallel downloads, dependency solving, and reproducible installs.
📦 pak.r-lib.org
#RStats
The {pak} package speeds things up with parallel downloads, dependency solving, and reproducible installs.
📦 pak.r-lib.org
#RStats

September 23, 2025 at 2:22 PM
Ever run `install.packages()` and wish it were faster, smarter, and more reliable?
The {pak} package speeds things up with parallel downloads, dependency solving, and reproducible installs.
📦 pak.r-lib.org
#RStats
The {pak} package speeds things up with parallel downloads, dependency solving, and reproducible installs.
📦 pak.r-lib.org
#RStats
Come and work with Nicole Stoesser and I in Oxford with the fantastic team @modmedmicro.bsky.social - great opportunity for a postdoc in microbial genomics to do some creative research with great datasets as part of our HPRU.
my.corehr.com/pls/uoxrecru...
my.corehr.com/pls/uoxrecru...
Job Details
my.corehr.com
September 23, 2025 at 9:47 AM
Come and work with Nicole Stoesser and I in Oxford with the fantastic team @modmedmicro.bsky.social - great opportunity for a postdoc in microbial genomics to do some creative research with great datasets as part of our HPRU.
my.corehr.com/pls/uoxrecru...
my.corehr.com/pls/uoxrecru...
Reposted by Sam Lipworth
Identification of specific metabolic capacities associated with major extraintestinal pathogenic Escherichia coli lineages https://www.biorxiv.org/content/10.1101/2025.09.14.675865v1
September 15, 2025 at 3:34 AM
Identification of specific metabolic capacities associated with major extraintestinal pathogenic Escherichia coli lineages https://www.biorxiv.org/content/10.1101/2025.09.14.675865v1
Great new work from #DotNagy @modmedmicro.bsky.social - she tool 96 clinical isolates from the NEKSUS study and sequenced with both Illumina and Nanopore - question was, can we do away with hybrid sequencing for the rest of the study?
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

Nanopore long-read only genome assembly of clinical Enterobacterales isolates is complete and accurate
Whole bacterial genome sequence reconstruction using Oxford Nanopore Technologies ('Nanopore') long-read only sequencing may offer a lower-cost, higher-throughput alternative for pathogen surveillance...
www.biorxiv.org
September 18, 2025 at 11:38 AM
Great new work from #DotNagy @modmedmicro.bsky.social - she tool 96 clinical isolates from the NEKSUS study and sequenced with both Illumina and Nanopore - question was, can we do away with hybrid sequencing for the rest of the study?
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Reposted by Sam Lipworth
Sometimes you meet absolutely incredible bioinfo-magicians.
It was a huge privilege when @shenwei356.bsky.social
joined our group for a year on an @embl.org sabbatical.
While here, he developed a new way of aligning to
millions of bacteria, called LexicMap 1/n
www.nature.com/articles/s41...
It was a huge privilege when @shenwei356.bsky.social
joined our group for a year on an @embl.org sabbatical.
While here, he developed a new way of aligning to
millions of bacteria, called LexicMap 1/n
www.nature.com/articles/s41...

Efficient sequence alignment against millions of prokaryotic genomes with LexicMap - Nature Biotechnology
LexicMap uses a fixed set of probes to efficiently query gene sequences for fast and low-memory alignment.
www.nature.com
September 10, 2025 at 9:12 AM
Sometimes you meet absolutely incredible bioinfo-magicians.
It was a huge privilege when @shenwei356.bsky.social
joined our group for a year on an @embl.org sabbatical.
While here, he developed a new way of aligning to
millions of bacteria, called LexicMap 1/n
www.nature.com/articles/s41...
It was a huge privilege when @shenwei356.bsky.social
joined our group for a year on an @embl.org sabbatical.
While here, he developed a new way of aligning to
millions of bacteria, called LexicMap 1/n
www.nature.com/articles/s41...
Reposted by Sam Lipworth
Very happy to be a (small) part of this story!
The role of plasmid-encoded ISs in the acceleration of AMR evolution, through inactivation of different bacterial genes.
The role of plasmid-encoded ISs in the acceleration of AMR evolution, through inactivation of different bacterial genes.
New paper out! 🔈🔈📣📣
Plasmids promote antimicrobial resistance through Insertion Sequence-mediated gene inactivation.
Combining experimental and computational approaches, we unveil how two of the most prevalent bacterial MGE accelerate the evolution of AMR. 🧵👇🏻
www.biorxiv.org/content/10.1...
Plasmids promote antimicrobial resistance through Insertion Sequence-mediated gene inactivation.
Combining experimental and computational approaches, we unveil how two of the most prevalent bacterial MGE accelerate the evolution of AMR. 🧵👇🏻
www.biorxiv.org/content/10.1...

Plasmids promote antimicrobial resistance through Insertion Sequence-mediated gene inactivation
Antimicrobial Resistance (AMR) is a major threat to public health. Plasmids are mobile genetic elements that can rapidly spread across bacterial populations, promoting the dissemination of AMR genes i...
www.biorxiv.org
August 13, 2025 at 11:09 AM
Very happy to be a (small) part of this story!
The role of plasmid-encoded ISs in the acceleration of AMR evolution, through inactivation of different bacterial genes.
The role of plasmid-encoded ISs in the acceleration of AMR evolution, through inactivation of different bacterial genes.
Reposted by Sam Lipworth
New analysis from Kevin Chau @modmedmicro.bsky.social for the SinkBug Consortium (a collaboration with teams across 29 UK hospitals, NITCAR, UKHSA): Pathogen and AMR gene burden in 287 hospital sink-trap metagenomes and factors associated with high burdens.
The hospital Sink-ome: Pathogen and antimicrobial resistance gene burden in sink-traps across 29 UK hospitals and associations with sink characteristics https://www.medrxiv.org/content/10.1101/2025.07.25.25332191v1
July 26, 2025 at 9:47 AM
New analysis from Kevin Chau @modmedmicro.bsky.social for the SinkBug Consortium (a collaboration with teams across 29 UK hospitals, NITCAR, UKHSA): Pathogen and AMR gene burden in 287 hospital sink-trap metagenomes and factors associated with high burdens.
Reposted by Sam Lipworth
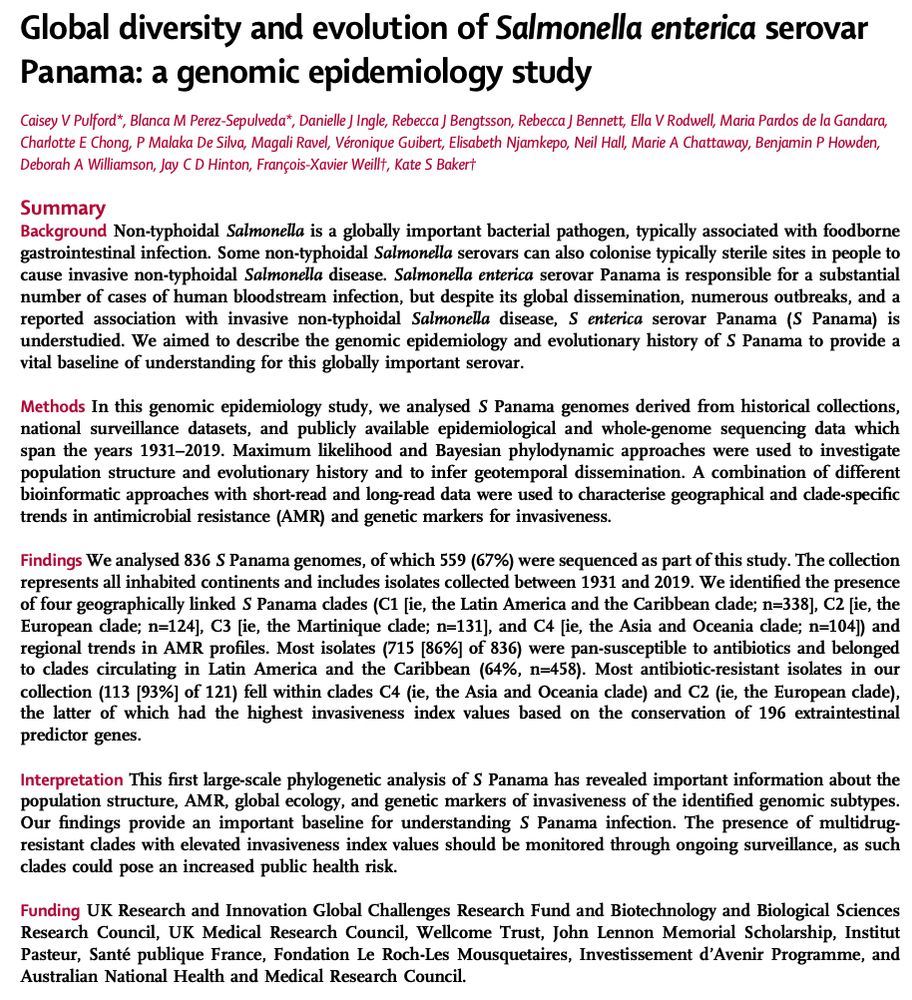
There are half a million cases of invasive non-Typhoidal Salmonella (iNTS) every year and while you might know S. Typhimurim and Enteritidis, you may not have heard of S. Panama 💩 🩸 🧫 💊 Check out our latest in @lancetmicrobe.bsky.social doi.org/10.1016/j.la... and the 🧵 below to find out more 1/n
July 27, 2025 at 8:35 AM
There are half a million cases of invasive non-Typhoidal Salmonella (iNTS) every year and while you might know S. Typhimurim and Enteritidis, you may not have heard of S. Panama 💩 🩸 🧫 💊 Check out our latest in @lancetmicrobe.bsky.social doi.org/10.1016/j.la... and the 🧵 below to find out more 1/n
Reposted by Sam Lipworth
BEAST X v10.5.0 finally released – you can't just do beta releases forever. github.com/beast-dev/be...
Details and instructions for installing on the BEAST website: beast.community
Details and instructions for installing on the BEAST website: beast.community

July 2, 2025 at 8:32 PM
BEAST X v10.5.0 finally released – you can't just do beta releases forever. github.com/beast-dev/be...
Details and instructions for installing on the BEAST website: beast.community
Details and instructions for installing on the BEAST website: beast.community
Reposted by Sam Lipworth
Delighted to see this paper from danderson123.bsky.social 's PhD out. We have been building tools for AMR gene detection for over a decade now, but multicopy genes remain challenging. Dan shows that with a gene-space de Bruijn graph and long reads, you can do well
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
May 19, 2025 at 9:28 AM
Delighted to see this paper from danderson123.bsky.social 's PhD out. We have been building tools for AMR gene detection for over a decade now, but multicopy genes remain challenging. Dan shows that with a gene-space de Bruijn graph and long reads, you can do well
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Reposted by Sam Lipworth
Important paper by Lance Thurlow, @bconlon.bsky.social @sirmicrobe.bsky.social and others:
Diabetes potentiates the emergence and expansion of antibiotic resistance.
"hyperglycemia..facilitates.expansion and takeover of resistant mutants in diabetic infections."
www.science.org/doi/10.1126/...
Diabetes potentiates the emergence and expansion of antibiotic resistance.
"hyperglycemia..facilitates.expansion and takeover of resistant mutants in diabetic infections."
www.science.org/doi/10.1126/...

Diabetes potentiates the emergence and expansion of antibiotic resistance
Diabetic infections are a reservoir for the emergence and proliferation of antibiotic resistance in Staphylococcus aureus.
www.science.org
February 14, 2025 at 8:24 PM
Important paper by Lance Thurlow, @bconlon.bsky.social @sirmicrobe.bsky.social and others:
Diabetes potentiates the emergence and expansion of antibiotic resistance.
"hyperglycemia..facilitates.expansion and takeover of resistant mutants in diabetic infections."
www.science.org/doi/10.1126/...
Diabetes potentiates the emergence and expansion of antibiotic resistance.
"hyperglycemia..facilitates.expansion and takeover of resistant mutants in diabetic infections."
www.science.org/doi/10.1126/...
Reposted by Sam Lipworth
Antimicrobial resistance data is now on the UKHSA data dashboard! 🧫
ukhsa-dashboard.data.gov.uk/antimicrobia...
ukhsa-dashboard.data.gov.uk/antimicrobia...

February 10, 2025 at 7:43 PM
Antimicrobial resistance data is now on the UKHSA data dashboard! 🧫
ukhsa-dashboard.data.gov.uk/antimicrobia...
ukhsa-dashboard.data.gov.uk/antimicrobia...
Reposted by Sam Lipworth
New research article in The Lancet Microbe
Prevalence, misclassification, and clinical consequences of the heteroresistant phenotype in Escherichia coli bloodstream infections in patients in Uppsala, Sweden: a retrospective cohort study
www.thelancet.com/journals/lan...
#IDSky #ClinMicro #AMR
Prevalence, misclassification, and clinical consequences of the heteroresistant phenotype in Escherichia coli bloodstream infections in patients in Uppsala, Sweden: a retrospective cohort study
www.thelancet.com/journals/lan...
#IDSky #ClinMicro #AMR

January 16, 2025 at 4:02 PM
New research article in The Lancet Microbe
Prevalence, misclassification, and clinical consequences of the heteroresistant phenotype in Escherichia coli bloodstream infections in patients in Uppsala, Sweden: a retrospective cohort study
www.thelancet.com/journals/lan...
#IDSky #ClinMicro #AMR
Prevalence, misclassification, and clinical consequences of the heteroresistant phenotype in Escherichia coli bloodstream infections in patients in Uppsala, Sweden: a retrospective cohort study
www.thelancet.com/journals/lan...
#IDSky #ClinMicro #AMR

