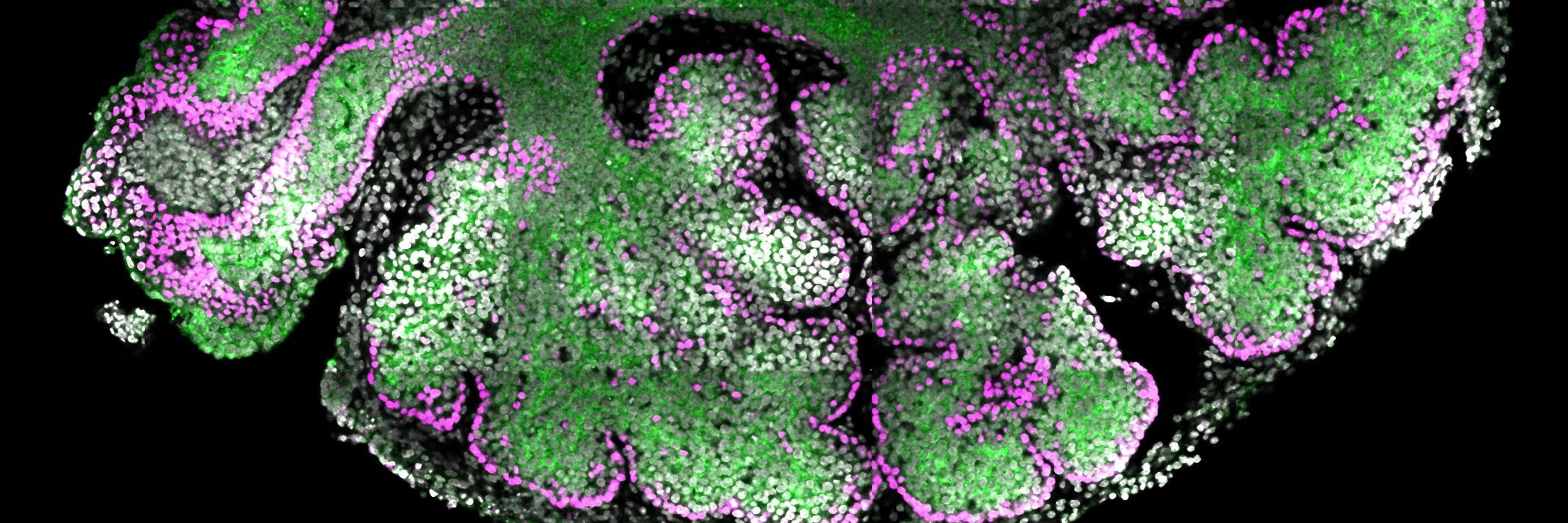
Tissue morphogenesis & Fate specification in epithelia.
Organoid and live imaging.
Alumni AgroParisTech & ENS Ulm
We built a single-cell atlas of 14 multilayered epithelia and revealed a conserved transcriptomic program guiding tissue architecture and fate composition. Our work brings decades of tissue-specific studies together into a unified evo-devo framework.
www.biorxiv.org/content/10.1...
Thanks to @frelab.bsky.social , the lab, and all collaborators involved.
Special thanks to @candice-merle.bsky.social for her help throughout the project.
genetics.org.uk/medals-and-p...

Thanks to @frelab.bsky.social , the lab, and all collaborators involved.
Special thanks to @candice-merle.bsky.social for her help throughout the project.



www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
#barcodes #plasticity
#barcodes #plasticity
Cross-organ (14 in🐭), Cross-species (7🪰🐟x2🐸🐥🐭👤) single-cell transcriptomics
p63+Jag2+ basal signal sendor▶️ p63-Hes1+ suprabasal receivers🐭
bioRxiv 2025
www.biorxiv.org/content/10.1...

Cross-organ (14 in🐭), Cross-species (7🪰🐟x2🐸🐥🐭👤) single-cell transcriptomics
p63+Jag2+ basal signal sendor▶️ p63-Hes1+ suprabasal receivers🐭
bioRxiv 2025
www.biorxiv.org/content/10.1...
We built a single-cell atlas of 14 multilayered epithelia and revealed a conserved transcriptomic program guiding tissue architecture and fate composition. Our work brings decades of tissue-specific studies together into a unified evo-devo framework.
www.biorxiv.org/content/10.1...
We built a single-cell atlas of 14 multilayered epithelia and revealed a conserved transcriptomic program guiding tissue architecture and fate composition. Our work brings decades of tissue-specific studies together into a unified evo-devo framework.
www.biorxiv.org/content/10.1...
www.cell.com/developmenta...

www.cell.com/developmenta...
doi.org/10.1016/j.de...

doi.org/10.1016/j.de...

Now out: rdcu.be/eIIKD
A great contribution from Jakub Sumbal, showing how stromal cells, and diverse fibroblast subsets, regulate branching morphogenesis.
@sumbalovakoledova.bsky.social @frelab.bsky.social

Now out: rdcu.be/eIIKD
A great contribution from Jakub Sumbal, showing how stromal cells, and diverse fibroblast subsets, regulate branching morphogenesis.
@sumbalovakoledova.bsky.social @frelab.bsky.social
training.institut-curie.org/courses/deve...
training.institut-curie.org/courses/deve...
We reveal how 4 branched epithelia—mammary, lacrimal, salivary & prostate—use a conserved YAP–Notch–p63 circuit to self-organize during development & regeneration.
Here’s the story👇
authors.elsevier.com/c/1lM285Sx5g...
Work done in @frelab.bsky.social at
@institutcurie.bsky.social
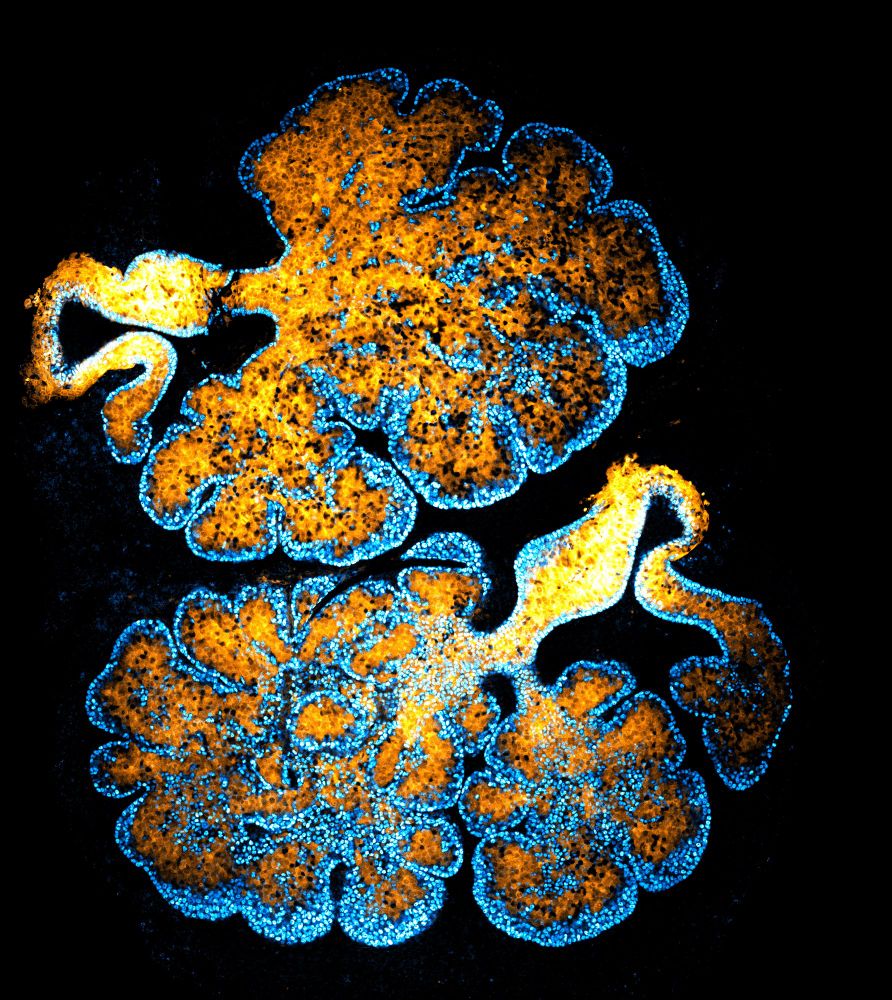
We reveal how 4 branched epithelia—mammary, lacrimal, salivary & prostate—use a conserved YAP–Notch–p63 circuit to self-organize during development & regeneration.
Here’s the story👇
authors.elsevier.com/c/1lM285Sx5g...
Work done in @frelab.bsky.social at
@institutcurie.bsky.social
Silvia Fre and coworkers @frelab.bsky.social @institutcurie.bsky.social
www.embopress.org/doi/full/10....

Silvia Fre and coworkers @frelab.bsky.social @institutcurie.bsky.social
www.embopress.org/doi/full/10....
@ehannezo.bsky.social and all the co-authors!

@ehannezo.bsky.social and all the co-authors!
, Kevan Shokat, and I are excited to announce a historic achievement by our team at @vevotherapeutics.bsky.social: tinyurl.com/4rrvap3t
, Kevan Shokat, and I are excited to announce a historic achievement by our team at @vevotherapeutics.bsky.social: tinyurl.com/4rrvap3t
Our #Stemcell #DevBio and #Regeneration
story by Silvia Fre's lab at InstitutCurie is on biorXiv.
Link: tinyurl.com/2uspcd3u
We reveal how 4 epithelia self-organize during embryonic development and regeneration.
Our #Stemcell #DevBio and #Regeneration
story by Silvia Fre's lab at InstitutCurie is on biorXiv.
Link: tinyurl.com/2uspcd3u
We reveal how 4 epithelia self-organize during embryonic development and regeneration.

