Petr Šulc
@petrsulc.bsky.social
Associate Professor at Arizona State University, and ERC grant PI at TU Munich. We play with DNA and RNA to make nanoscale structures and devices. Find out more at our lab page: sulclab.org
Reposted by Petr Šulc
Thank you @mskcancercenter.bsky.social for highlighting our work on #viralmimicry in our genome, its evolution, the selective forces on it, and its potential role in cancer.
A great years long collaborative effort with @petrsulc.bsky.social, @daniel-decarvalho.bsky.social, & many great colleagues!
A great years long collaborative effort with @petrsulc.bsky.social, @daniel-decarvalho.bsky.social, & many great colleagues!
Dr. @bengrbm.bsky.social has tried to shed light on the innate immune system and how it affects cancer cells as they evolve.
Now Dr. Greenbaum’s lab has been taking a deep dive into modeling viral mimicry, trying to understand how this could shape cancer cells’ development process. bit.ly/3VWadA8
Now Dr. Greenbaum’s lab has been taking a deep dive into modeling viral mimicry, trying to understand how this could shape cancer cells’ development process. bit.ly/3VWadA8

October 8, 2025 at 11:56 PM
Thank you @mskcancercenter.bsky.social for highlighting our work on #viralmimicry in our genome, its evolution, the selective forces on it, and its potential role in cancer.
A great years long collaborative effort with @petrsulc.bsky.social, @daniel-decarvalho.bsky.social, & many great colleagues!
A great years long collaborative effort with @petrsulc.bsky.social, @daniel-decarvalho.bsky.social, & many great colleagues!
🧪 In our new work (just published in Cell Genomics) with Benjamin Greenbaum , John LaCava, Daniel De Carvalho, PhD , Simona Cocco and Remi Monasson labs, we venture into the dark matter of human genome:
www.cell.com/cell-genomic...
🧵A shot thread summary below in comments:⬇️
www.cell.com/cell-genomic...
🧵A shot thread summary below in comments:⬇️

Repeats mimic pathogen-associated patterns across a vast evolutionary landscape
An emerging hallmark of disease is transcription of pathogen-associated molecular
patterns from within the genome–known as viral mimicry. We propose a statistical physics
framework to measure “selecti...
www.cell.com
October 4, 2025 at 9:49 PM
🧪 In our new work (just published in Cell Genomics) with Benjamin Greenbaum , John LaCava, Daniel De Carvalho, PhD , Simona Cocco and Remi Monasson labs, we venture into the dark matter of human genome:
www.cell.com/cell-genomic...
🧵A shot thread summary below in comments:⬇️
www.cell.com/cell-genomic...
🧵A shot thread summary below in comments:⬇️
Reposted by Petr Šulc
On this first day of #DNA31, reminder for the mol pro community that I put together this feed which picks up the last 3 days of posts from our community! Enjoy the meeting! 🧬
Are there any other scientists from molecular programming here? That's self-assembly, chemical computation, rational design et al. I made a mol pro feed which works like the Science (🧪) feed. Let me know, I add you to the list, you post with a 🧬 emoji. Let's assemble!
bsky.app/profile/flop...
bsky.app/profile/flop...
August 25, 2025 at 8:12 AM
On this first day of #DNA31, reminder for the mol pro community that I put together this feed which picks up the last 3 days of posts from our community! Enjoy the meeting! 🧬
A great effort to chart possible future paths for the molecular programming field
Podcast drop🧬! As part of the Molecular Programming Flightplan, we assembled a panel discussion on collaboration! The panel is now available as two podcasts, each with half the panel, and a bonus interview with @programmablematter.bsky.social.
Links to all major feeds here:
podcast.molpi.gs
Links to all major feeds here:
podcast.molpi.gs
July 17, 2025 at 7:43 AM
A great effort to chart possible future paths for the molecular programming field
🧪🧬
Our new paper is just out in PNAS:
www.pnas.org/doi/10.1073/...
It is about nucleic acid strand displacement, which is a key reaction in molecular computation, and part of the success of the oxDNA model comes from the fact it can efficiently simulate this process.
1/4🧵
Our new paper is just out in PNAS:
www.pnas.org/doi/10.1073/...
It is about nucleic acid strand displacement, which is a key reaction in molecular computation, and part of the success of the oxDNA model comes from the fact it can efficiently simulate this process.
1/4🧵
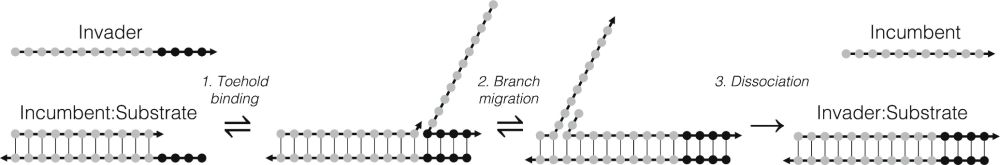
Controlling DNA–RNA strand displacement kinetics with base distribution | PNAS
DNA–RNA hybrid strand displacement underpins the function of many natural and engineered
systems. Understanding and controlling factors affecting D...
www.pnas.org
June 7, 2025 at 11:05 AM
🧪🧬
Our new paper is just out in PNAS:
www.pnas.org/doi/10.1073/...
It is about nucleic acid strand displacement, which is a key reaction in molecular computation, and part of the success of the oxDNA model comes from the fact it can efficiently simulate this process.
1/4🧵
Our new paper is just out in PNAS:
www.pnas.org/doi/10.1073/...
It is about nucleic acid strand displacement, which is a key reaction in molecular computation, and part of the success of the oxDNA model comes from the fact it can efficiently simulate this process.
1/4🧵
Just in time for #FNANO conference in Snowbird, UT, our updated and improved version of nanobase.org server is online. Just like the PDB database helped to share protein structures, we want to create a community resource for the DNA/RNA/protein nanotechnology design researchers to share their work
Nanobase
A repository for DNA/RNA nanotechnology
nanobase.org
April 26, 2025 at 10:23 PM
Just in time for #FNANO conference in Snowbird, UT, our updated and improved version of nanobase.org server is online. Just like the PDB database helped to share protein structures, we want to create a community resource for the DNA/RNA/protein nanotechnology design researchers to share their work
🧪In our new paper (journals.aps.org/prl/abstract...) in collaboration with Russo, Romano, Rovigatti and Sciortino groups in Rome / Venice, we look at Classical Nucleation Theory: a popular model of nucleation process, n is a key phenomena in self-assembly, self-organization and phase transitions.
Falsifiability Test for Classical Nucleation Theory
Classical nucleation theory (CNT) is built upon the capillarity approximation, i.e., the assumption that the nucleation properties can be inferred from the bulk properties of the melt and the crystal....
journals.aps.org
April 8, 2025 at 6:00 PM
🧪In our new paper (journals.aps.org/prl/abstract...) in collaboration with Russo, Romano, Rovigatti and Sciortino groups in Rome / Venice, we look at Classical Nucleation Theory: a popular model of nucleation process, n is a key phenomena in self-assembly, self-organization and phase transitions.
Great collaboration with @laklab-tubs.bsky.social , simulations carried out by my student Josh Evans
Very excited to introduce mismatch-assisted toehold exchange (MATE) concept for the first time, our new magnetic diagnostics cascades:
biorxiv.org/content/10.1...
Excellent work by Rebecca, "Metrology4life" 2nd PhD student and Great collaboration with @petrsulc.bsky.social
biorxiv.org/content/10.1...
Excellent work by Rebecca, "Metrology4life" 2nd PhD student and Great collaboration with @petrsulc.bsky.social

Mismatch-Assisted Toehold Exchange Cascades for Magnetic Nanoparticle-Based Nucleic Acid Diagnostics
Sensitive, simple, and rapid detection of nucleic acid sequences at point-of-care (POC) settings is still an unmet quest. Magnetic readout assays combined with toehold-mediated strand displacement (TM...
biorxiv.org
March 17, 2025 at 12:42 PM
Great collaboration with @laklab-tubs.bsky.social , simulations carried out by my student Josh Evans
🧬 A new tutorial article from our student Sarah, walking step by step through DNA origami simulation using oxDNA model, with the help of oxview.org and oxdna.org free tools. No prior experience assumed, and also accompanied by a video: youtu.be/5-rgMekX8gE
onlinelibrary.wiley.com/doi/10.1002/...
onlinelibrary.wiley.com/doi/10.1002/...
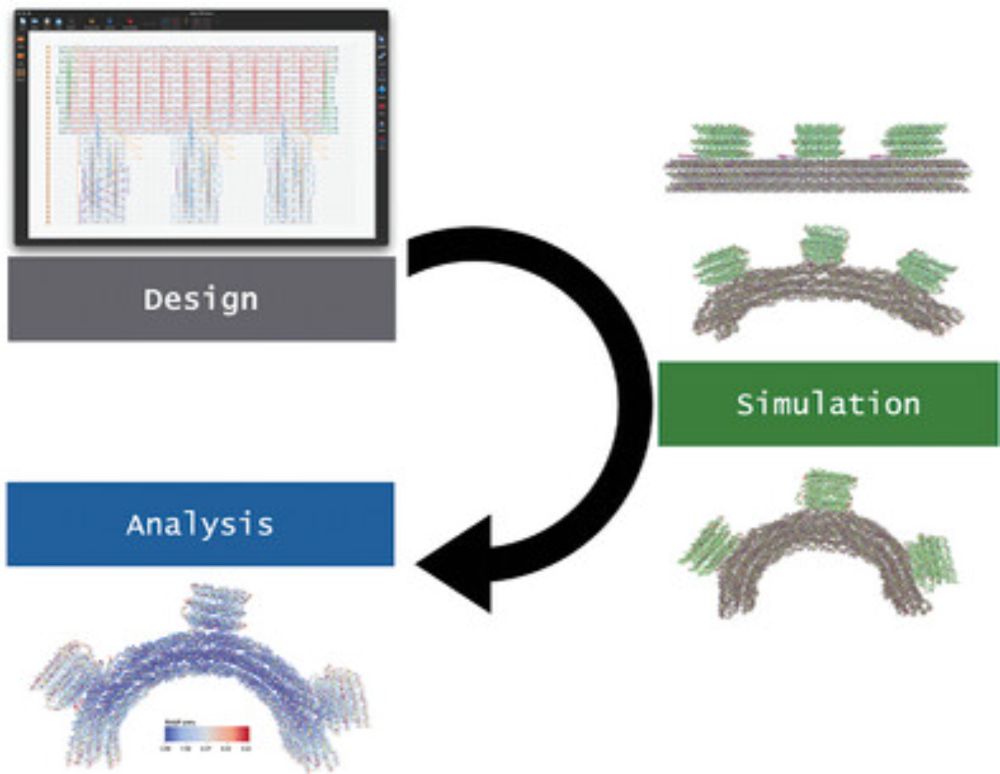
How We Simulate DNA Origami
This tutorial provides a detailed step-by-step description on how to import DNA origami designs into the oxDNA model and characterize the origami's properties (such as equilibrium shape and flexibili...
onlinelibrary.wiley.com
February 5, 2025 at 10:57 AM
🧬 A new tutorial article from our student Sarah, walking step by step through DNA origami simulation using oxDNA model, with the help of oxview.org and oxdna.org free tools. No prior experience assumed, and also accompanied by a video: youtu.be/5-rgMekX8gE
onlinelibrary.wiley.com/doi/10.1002/...
onlinelibrary.wiley.com/doi/10.1002/...
I am very grateful to Neuron Foundation, considered among the most prestigious awards for Czech Scientists, for awarding me Promising Scientist award in Physics yesterday at a ceremony in Prague. Many thanks goes to my family, collaborators and students: www.nadaceneuron.cz/novinky/ceny...

Ceny Neuron 2024: Seznamte se s letošními laureáty | Nadace Neuron
Věda je plná emocí a práce vědce představuje rébus, hru, touhu po poznání, vášeň, radost i lásku. Prozkoumejte krásu těchto emocí prostřednictvím laureátů Ceny Neuron.
www.nadaceneuron.cz
January 20, 2025 at 10:03 AM
I am very grateful to Neuron Foundation, considered among the most prestigious awards for Czech Scientists, for awarding me Promising Scientist award in Physics yesterday at a ceremony in Prague. Many thanks goes to my family, collaborators and students: www.nadaceneuron.cz/novinky/ceny...
🧪🧬 New article with J Russo, D. Pinto and F. Sciortino out is @acspublications.bsky.social ACS Nano: pubs.acs.org/doi/10.1021/...
We use our #SAT Assembly method to design particles that assemble into 3D quasicrystal clusters, paving a way to their experimental realization with DNA nanostructures
We use our #SAT Assembly method to design particles that assemble into 3D quasicrystal clusters, paving a way to their experimental realization with DNA nanostructures
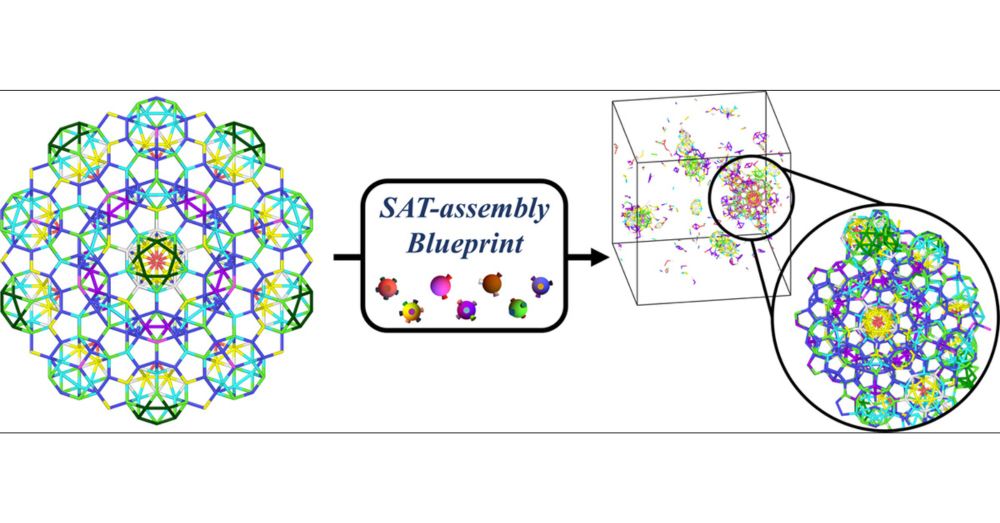
Automating Blueprints for the Assembly of Colloidal Quasicrystal Clusters
One of the frontiers of nanotechnology is advancing beyond the periodic self-assembly of materials. Icosahedral quasicrystals, aperiodic in all directions, represent one of the most challenging target...
pubs.acs.org
December 22, 2024 at 8:48 AM
🧪🧬 New article with J Russo, D. Pinto and F. Sciortino out is @acspublications.bsky.social ACS Nano: pubs.acs.org/doi/10.1021/...
We use our #SAT Assembly method to design particles that assemble into 3D quasicrystal clusters, paving a way to their experimental realization with DNA nanostructures
We use our #SAT Assembly method to design particles that assemble into 3D quasicrystal clusters, paving a way to their experimental realization with DNA nanostructures
In our Christmas tradition with John Russo's lab at Sapienza university of Rome, here is a Christmas-themed video highlighting a current research project based on our #SAT-assembly method, this time chiral crystals self-assembly (made by C. Beneduce): www.youtube.com/watch?v=0A7x...

SATa CLAUSE: When magic snow falls, a enchanted meadow blossoms
YouTube video by Sapienza soft matter group
www.youtube.com
December 13, 2024 at 1:13 AM
In our Christmas tradition with John Russo's lab at Sapienza university of Rome, here is a Christmas-themed video highlighting a current research project based on our #SAT-assembly method, this time chiral crystals self-assembly (made by C. Beneduce): www.youtube.com/watch?v=0A7x...
Reposted by Petr Šulc
Thank you so much to whoever put this very comprehensive list of #science Bsky starter packs together. This is just amazing: docs.google.com/document/d/1...
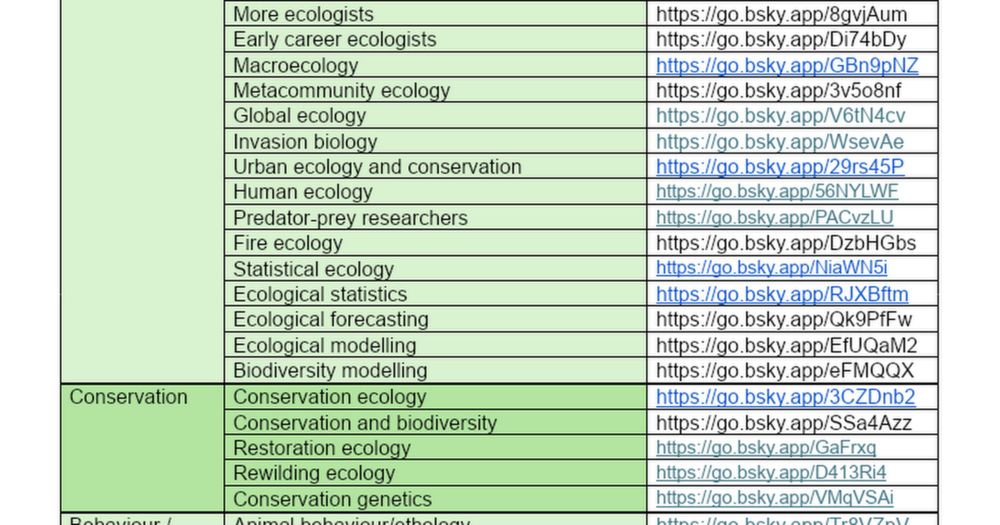
Starter packs
BIOLOGY General Science community https://go.bsky.app/HVYAMEA Research Institutions in Life Sciences https://go.bsky.app/LxXpcvJ LGBTQIA+ in STEM https://go.bsky.app/BFc4wgf LGBTQIA+ in STEM 2 h...
docs.google.com
November 26, 2024 at 2:47 AM
Thank you so much to whoever put this very comprehensive list of #science Bsky starter packs together. This is just amazing: docs.google.com/document/d/1...
Are you working in DNA/RNA nanotech? Add your name to the list!
To accelerate the X-odus, I created a DNA & RNA nanotech starter pack :) Comment if you want to be added!
go.bsky.app/7LUeJ61
go.bsky.app/7LUeJ61
November 25, 2024 at 8:07 PM
Are you working in DNA/RNA nanotech? Add your name to the list!
Our new article on combining ML with experiments to design peptide binders to a target proteins is just out in ACS Central Science: pubs.acs.org/doi/10.1021/... .
We show it is possible to iteratively combine training on small dataset and experiments to achieve higher binding affinity.
We show it is possible to iteratively combine training on small dataset and experiments to achieve higher binding affinity.
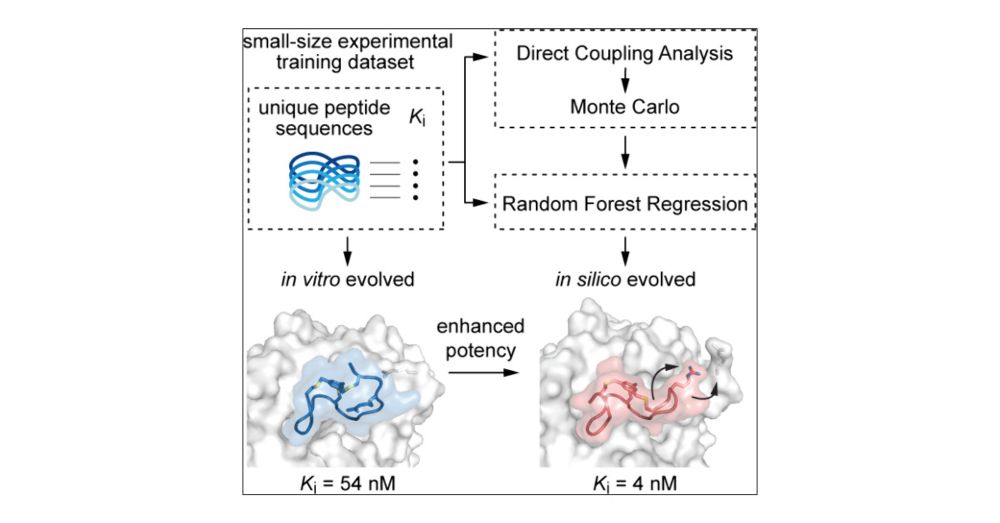
Combination of Coevolutionary Information and Supervised Learning Enables Generation of Cyclic Peptide Inhibitors with Enhanced Potency from a Small Data Set
Computational generation of cyclic peptide inhibitors using machine learning models requires large size training data sets often difficult to generate experimentally. Here we demonstrated that sequent...
pubs.acs.org
November 20, 2024 at 6:16 PM
Our new article on combining ML with experiments to design peptide binders to a target proteins is just out in ACS Central Science: pubs.acs.org/doi/10.1021/... .
We show it is possible to iteratively combine training on small dataset and experiments to achieve higher binding affinity.
We show it is possible to iteratively combine training on small dataset and experiments to achieve higher binding affinity.
🧪 🧬 Can code be evolved? In our collaboration with S. Forrest group, we show that random "mutations" to the code, accepted if they pass unit tests, can actually find changes to CUDA version of oxDNA simulation tool for DNA nanotechnology that lead to a speedup:
doi.org/10.1145/3703...
doi.org/10.1145/3703...

Evolving to find optimizations humans miss: using evolutionary computation to improve GPU code for bioinformatics applications | ACM Transactions on Evolutionary Learning and Optimization
GPUs are used in many settings to accelerate large-scale scientific computation, including
simulation, computational biology, and molecular dynamics. However, optimizing codes
to run efficiently on GP...
doi.org
November 17, 2024 at 12:33 AM
🧪 🧬 Can code be evolved? In our collaboration with S. Forrest group, we show that random "mutations" to the code, accepted if they pass unit tests, can actually find changes to CUDA version of oxDNA simulation tool for DNA nanotechnology that lead to a speedup:
doi.org/10.1145/3703...
doi.org/10.1145/3703...
Reposted by Petr Šulc
First up, Ned Seeman's 1982 "Nucleic Acid Junctions and Lattices", published in the Journal of Theoretical Biology. This was the first paper to propose that through sequence design it might be possible to create precisely-defined crystal lattices out of DNA. 🧬
www.sciencedirect.com/science/arti...
www.sciencedirect.com/science/arti...
November 12, 2024 at 10:01 PM
First up, Ned Seeman's 1982 "Nucleic Acid Junctions and Lattices", published in the Journal of Theoretical Biology. This was the first paper to propose that through sequence design it might be possible to create precisely-defined crystal lattices out of DNA. 🧬
www.sciencedirect.com/science/arti...
www.sciencedirect.com/science/arti...

