Petr Šulc
@petrsulc.bsky.social
Associate Professor at Arizona State University, and ERC grant PI at TU Munich. We play with DNA and RNA to make nanoscale structures and devices. Find out more at our lab page: sulclab.org
While this work presents a first step towards 'decoding the repeatome' in human genome, it also presents a fast an accessible framework to assess stimulatory potential of transcripts coming from various experimental datasets.
October 4, 2025 at 9:49 PM
While this work presents a first step towards 'decoding the repeatome' in human genome, it also presents a fast an accessible framework to assess stimulatory potential of transcripts coming from various experimental datasets.
Complementary to this hypothesis, it is also possible that maintaining these patterns is also beneficial for the repeat families, as we argue in the case of the highly studied family of LINE-1 repeats, where high CpG content might help prevent their deleterious effect on organism's fitness.
October 4, 2025 at 9:49 PM
Complementary to this hypothesis, it is also possible that maintaining these patterns is also beneficial for the repeat families, as we argue in the case of the highly studied family of LINE-1 repeats, where high CpG content might help prevent their deleterious effect on organism's fitness.
We argue that these repeats might have been co-opted to trigger innate immune system response upon translational dysregulation, such as in cancer cells. We checked experimentally that repeat transcripts that we marked as anomalous do bind to pattern recognition receptors of innate immune system.
October 4, 2025 at 9:49 PM
We argue that these repeats might have been co-opted to trigger innate immune system response upon translational dysregulation, such as in cancer cells. We checked experimentally that repeat transcripts that we marked as anomalous do bind to pattern recognition receptors of innate immune system.
It allows us to quantify how anomalous given sequence is with respect to what one would expect from a typical transcript from human genomes. The two chosen motifs, CpG content and double-stranded regions,are hallmarks of RNA viruses and our immune system recognizes them as immunostimulatory targets.
October 4, 2025 at 9:49 PM
It allows us to quantify how anomalous given sequence is with respect to what one would expect from a typical transcript from human genomes. The two chosen motifs, CpG content and double-stranded regions,are hallmarks of RNA viruses and our immune system recognizes them as immunostimulatory targets.
In our work, we instead focus on developing statistical mechanics-based framework to identify anomalous sequence (CpGf) and structure (long double-stranded segment) motifs in transcripts of these regions, which are known to be recognized by innate immune system receptors.
October 4, 2025 at 9:49 PM
In our work, we instead focus on developing statistical mechanics-based framework to identify anomalous sequence (CpGf) and structure (long double-stranded segment) motifs in transcripts of these regions, which are known to be recognized by innate immune system receptors.
A majority of our genome does not code for proteins nor has any established regulatory function. These regions are sometimes called 'junk DNA'. Their function, or lack of, is a very active topic of research.
October 4, 2025 at 9:49 PM
A majority of our genome does not code for proteins nor has any established regulatory function. These regions are sometimes called 'junk DNA'. Their function, or lack of, is a very active topic of research.
The predictions were verified experimentally. Our results have implications for DNA/RNA molecular computing, design of DNA computing systems that interface with RNA triggers, and as we show in our presented model, it can also have implications for predicting the kinetics in CRISPR-based systems.
June 7, 2025 at 11:05 AM
The predictions were verified experimentally. Our results have implications for DNA/RNA molecular computing, design of DNA computing systems that interface with RNA triggers, and as we show in our presented model, it can also have implications for predicting the kinetics in CRISPR-based systems.
It shows that just by permuting the distribution of bases in the duplex (while keeping AT / CG base pair number constant), the kinetics of the reaction can be altered by orders of magnitude, stemming from the details of differences in stability between hybrid and canonical bases in DNA/RNA systems.
June 7, 2025 at 11:05 AM
It shows that just by permuting the distribution of bases in the duplex (while keeping AT / CG base pair number constant), the kinetics of the reaction can be altered by orders of magnitude, stemming from the details of differences in stability between hybrid and canonical bases in DNA/RNA systems.
It is also of importance for biological systems like CRISPR-Cas9. In this collaboration, Eryk Ratajczyk supervised by Louis, Doye and Turberfield groups from Oxford, uses our new oxDNA-oxRNA hybrid coarse-grained model to study the strand displacement RNA invades DNA duplex.
June 7, 2025 at 11:05 AM
It is also of importance for biological systems like CRISPR-Cas9. In this collaboration, Eryk Ratajczyk supervised by Louis, Doye and Turberfield groups from Oxford, uses our new oxDNA-oxRNA hybrid coarse-grained model to study the strand displacement RNA invades DNA duplex.
Great work by our student Navraj and Subhajit, building on top of the ground work of previous database by @floppleton.bsky.social and Michael Matthies
April 26, 2025 at 10:24 PM
Great work by our student Navraj and Subhajit, building on top of the ground work of previous database by @floppleton.bsky.social and Michael Matthies
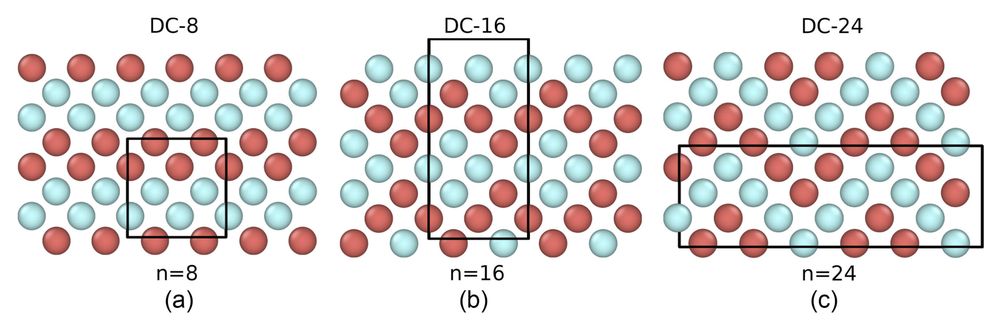
4/4: Our crystal example thus provides a falsifiability test for classical nucleation theory, showing need for also taking into account structural fluctuations.
Excellent work by Camilla Beneduce and Diogo Pinto from John Russo's group in Rome!
Excellent work by Camilla Beneduce and Diogo Pinto from John Russo's group in Rome!
April 8, 2025 at 6:00 PM
4/4: Our crystal example thus provides a falsifiability test for classical nucleation theory, showing need for also taking into account structural fluctuations.
Excellent work by Camilla Beneduce and Diogo Pinto from John Russo's group in Rome!
Excellent work by Camilla Beneduce and Diogo Pinto from John Russo's group in Rome!
3/4 However, we find that one of the three possible crystal phases is strongly preferred, due to the crystal lattice ordering being similar to the local ordering in the liquid phase.
April 8, 2025 at 6:00 PM
3/4 However, we find that one of the three possible crystal phases is strongly preferred, due to the crystal lattice ordering being similar to the local ordering in the liquid phase.
2/4: The assumptions of CNT rely on the free energy difference between bulk phase and the melt, and the interfacial free energy. In this work, we look at a two component system that forms a polymorph: it can nucleate into three different lattices with same interfacial and bulk free-energy.
April 8, 2025 at 6:00 PM
2/4: The assumptions of CNT rely on the free energy difference between bulk phase and the melt, and the interfacial free energy. In this work, we look at a two component system that forms a polymorph: it can nucleate into three different lattices with same interfacial and bulk free-energy.
Thank you Ulrich, it was a pleasure visiting Heidelberg and meeting you. Many thanks to the amazing group of @kgoepfrich.bsky.social for hosting me!
March 24, 2025 at 10:31 PM
Thank you Ulrich, it was a pleasure visiting Heidelberg and meeting you. Many thanks to the amazing group of @kgoepfrich.bsky.social for hosting me!
If you change something in the candano file, it has to be loaded again from scratch into oxview. The scadnano tool has recently been expanded to also allow for visualization in embedded oxview window. Having an interactive cadnano-like interface inside oxView is currently on our TBD wishlist
February 5, 2025 at 9:15 PM
If you change something in the candano file, it has to be loaded again from scratch into oxview. The scadnano tool has recently been expanded to also allow for visualization in embedded oxview window. Having an interactive cadnano-like interface inside oxView is currently on our TBD wishlist

