
https://pablo-arantes.github.io/
doi.org/10.26434/che...
ML has revolutionized protein modeling, but crucial challenges remain. For example, we can't reliably predict complicated protein structures without MSAs, which limits what we can design.
ML has revolutionized protein modeling, but crucial challenges remain. For example, we can't reliably predict complicated protein structures without MSAs, which limits what we can design.
We present eRMSF, a Python package for ensemble-based RMSF analysis of biomolecular systems, supporting MD, PDB, and ML ensembles.
Proud to collaborate with Rodrigo Ligabue-Braun and @conradopedebos.bsky.social
pubs.acs.org/doi/10.1021/...

We present eRMSF, a Python package for ensemble-based RMSF analysis of biomolecular systems, supporting MD, PDB, and ML ensembles.
Proud to collaborate with Rodrigo Ligabue-Braun and @conradopedebos.bsky.social
pubs.acs.org/doi/10.1021/...
Our paper is now published in JCIM!
Making ML-based force field parametrization open and easy for everyone.
With TorchANI, @rdkit.bsky.social & @openmm.org
🔗 pubs.acs.org/doi/abs/10.1...
@acs.org @giuliapalermo.bsky.social

Our paper is now published in JCIM!
Making ML-based force field parametrization open and easy for everyone.
With TorchANI, @rdkit.bsky.social & @openmm.org
🔗 pubs.acs.org/doi/abs/10.1...
@acs.org @giuliapalermo.bsky.social
Meet eRMSF, our new Python tool that tracks fluctuations over time!
Traditional RMSF shows “how much.”
eRMSF shows “when.” 🔥
Preprint 🔗 doi.org/10.26434/che...
GitHub github.com/pablo-arante...
Try it on Colab → colab.research.google.com/github/pablo...

Meet eRMSF, our new Python tool that tracks fluctuations over time!
Traditional RMSF shows “how much.”
eRMSF shows “when.” 🔥
Preprint 🔗 doi.org/10.26434/che...
GitHub github.com/pablo-arante...
Try it on Colab → colab.research.google.com/github/pablo...
www.nature.com/articles/s41...
www.nature.com/articles/s41...
People can easily refine parameters of multiple dihedral torsion for hundreds of molecules and employ those in FEP simulations.
10/10 recommend 👍
doi.org/10.26434/che...
People can easily refine parameters of multiple dihedral torsion for hundreds of molecules and employ those in FEP simulations.
10/10 recommend 👍
🚀 ParametrizANI - a #NeuralNetworks tool for molecular parametrization.
Predicts potential energy surfaces with near-DFT/CC accuracy, at a fraction of the computational cost!
#AI #QuantumChemistry #CompChem #ML 🤖
chemrxiv.org/engage/chemr...
Try it:
github.com/palermolab/P...

🚀 ParametrizANI - a #NeuralNetworks tool for molecular parametrization.
Predicts potential energy surfaces with near-DFT/CC accuracy, at a fraction of the computational cost!
#AI #QuantumChemistry #CompChem #ML 🤖
chemrxiv.org/engage/chemr...
Try it:
github.com/palermolab/P...
doi.org/10.26434/che...
doi.org/10.26434/che...
github.com/martinpacesa...
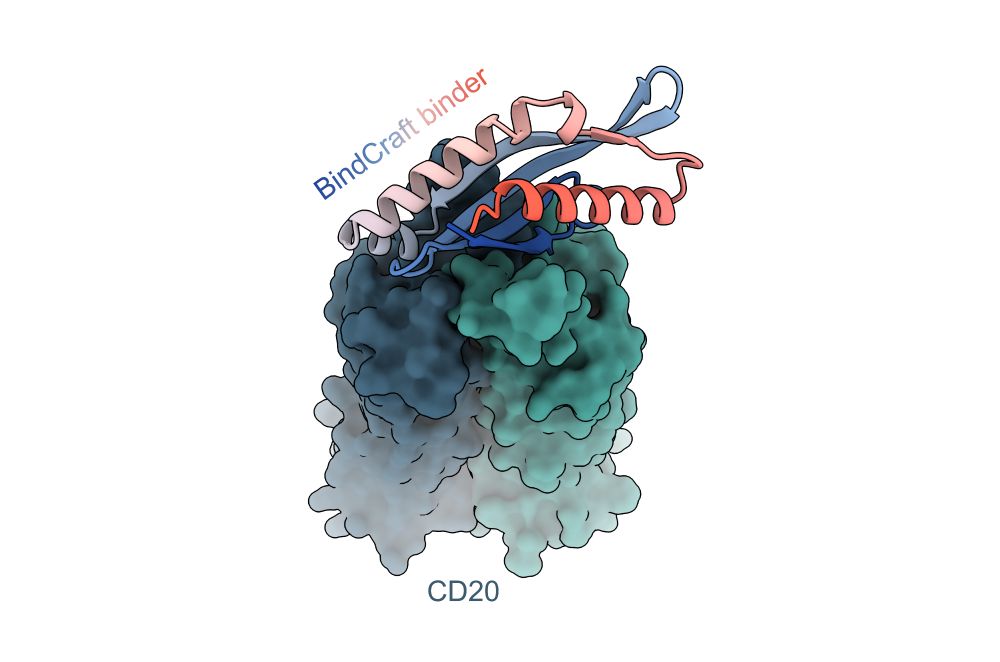
github.com/martinpacesa...
This is historic - insiders are blowing the whistle.
🧵(1/5)

This is historic - insiders are blowing the whistle.
🧵(1/5)
✨ Now with LogMD-based trajectory visualization.
🔗 Demo: rcsb.ai/ff9c2b1ee8
Feedback & collabs welcome! 🙌
🔗: GitHub: github.com/yehlincho/Bo...
🔗: Colab: colab.research.google.com/github/yehli...
@sokrypton.org @martinpacesa.bsky.social
✨ Now with LogMD-based trajectory visualization.
🔗 Demo: rcsb.ai/ff9c2b1ee8
Feedback & collabs welcome! 🙌
🔗: GitHub: github.com/yehlincho/Bo...
🔗: Colab: colab.research.google.com/github/yehli...
@sokrypton.org @martinpacesa.bsky.social
When someone takes a moment to say thank you, it reminds me why we keep pushing forward—sharing tools, writing posts, and trying to make science more open and accessible. Grateful for the kind words!

When someone takes a moment to say thank you, it reminds me why we keep pushing forward—sharing tools, writing posts, and trying to make science more open and accessible. Grateful for the kind words!
Turns out, CRISPR-associated transposons don’t just jump—they dance their way through DNA! 🕺🔬
Exciting times for genome engineering!
🧬 Read more in PNAS: www.pnas.org/doi/10.1073/...
#CRISPR #GeneEditing #PNAS #MDsimulations #CompChem
Turns out, CRISPR-associated transposons don’t just jump—they dance their way through DNA! 🕺🔬
Exciting times for genome engineering!
🧬 Read more in PNAS: www.pnas.org/doi/10.1073/...
#CRISPR #GeneEditing #PNAS #MDsimulations #CompChem
🔗 Try it on Google Colab: colab.research.google.com/github/pablo...
A huge thanks to Alexander Mathiasen for the support! 🙌
🔗 Try it on Google Colab: lnkd.in/gcuqd-fT
🔗 Try it on Google Colab: colab.research.google.com/github/pablo...
A huge thanks to Alexander Mathiasen for the support! 🙌
🔗 Try it on Google Colab: lnkd.in/gcuqd-fT
A huge thanks to Alexander Mathiasen for the support! 🙌
🔗 Try it on Google Colab: lnkd.in/gcuqd-fT
First, we made MD simulations rain down from the cloud. Now, we’re bringing deep learning into the mix! 🌩️🤖

First, we made MD simulations rain down from the cloud. Now, we’re bringing deep learning into the mix! 🌩️🤖


www.ks.uiuc.edu/Research/vmd...

www.ks.uiuc.edu/Research/vmd...



Audrey Lapinaite!
@narjournal.bsky.social
The dimerization of TadA8e and its exclusive interactions with #CRISPR Cas9 boost #BaseEditing efficiency!
academic.oup.com/nar/article/...
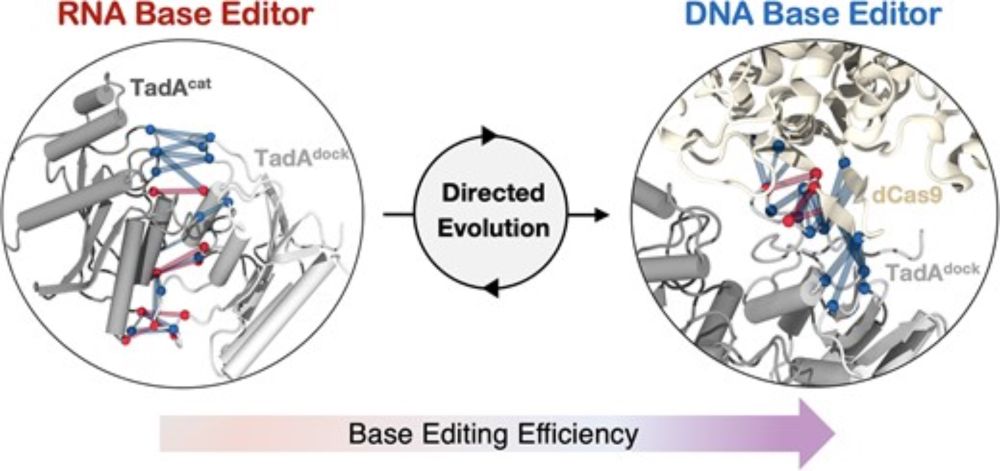
Audrey Lapinaite!
@narjournal.bsky.social
The dimerization of TadA8e and its exclusive interactions with #CRISPR Cas9 boost #BaseEditing efficiency!
academic.oup.com/nar/article/...
“Tú no puedes comprar al viento
Tú no puedes comprar al sol
Tú no puedes comprar la lluvia
Tú no puedes comprar el calor”

“Tú no puedes comprar al viento
Tú no puedes comprar al sol
Tú no puedes comprar la lluvia
Tú no puedes comprar el calor”
Historians: "that's a nazi salute"
Average person: "that's a nazi salute"
The Media: "Elon Musk makes odd gesture throwing his heart to the crowd."
Historians: "that's a nazi salute"
Average person: "that's a nazi salute"
The Media: "Elon Musk makes odd gesture throwing his heart to the crowd."

