
www.EpigenomeLab.com



academic.oup.com/bioinformati...
Published in @oxunipress.bsky.social Bioinformatics | led by aminnoorani.bsky.social
#Epigenomics #Genomics #Bioinformatics #MachineLearning #CUTandRUN #CompBio #4DNucleome
🔗 doi.org/10.1093/bioi...
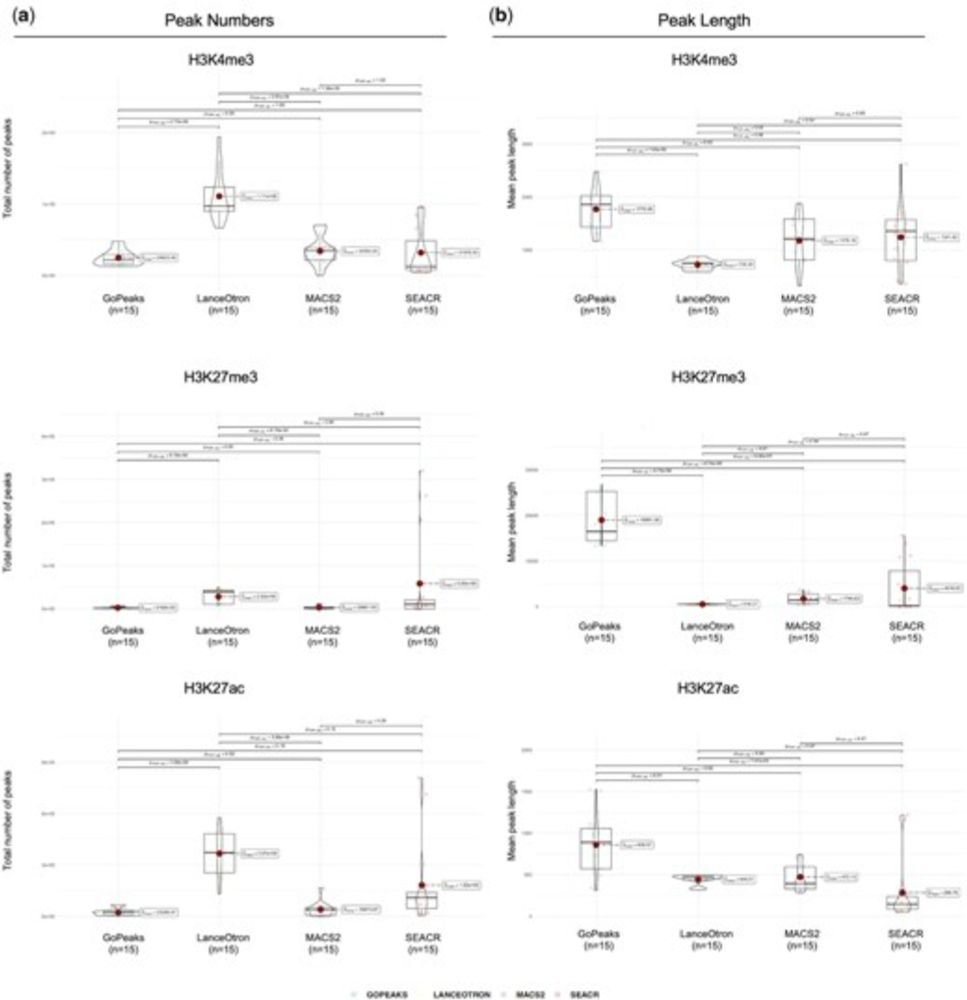
🔗 doi.org/10.1093/bioi...
academic.oup.com/bioinformati...
Published in @oxunipress.bsky.social Bioinformatics | led by aminnoorani.bsky.social
#Epigenomics #Genomics #Bioinformatics #MachineLearning #CUTandRUN #CompBio #4DNucleome



academic.oup.com/bioinformati...
Published in @oxunipress.bsky.social Bioinformatics | led by aminnoorani.bsky.social
#Epigenomics #Genomics #Bioinformatics #MachineLearning #CUTandRUN #CompBio #4DNucleome
ascopubs.org/doi/10.1200/...
Journal of Clinical Oncology @ascocancer.bsky.social
#CART #CellTherapy @ascopost.bsky.social @oncoalert.bsky.social @oncodaily.bsky.social

ascopubs.org/doi/10.1200/...
Journal of Clinical Oncology @ascocancer.bsky.social
#CART #CellTherapy @ascopost.bsky.social @oncoalert.bsky.social @oncodaily.bsky.social
Happy 2025 everyone 🎆 !

Happy 2025 everyone 🎆 !

