
Massive update of preprint with @polardlab.bsky.social!
Bacteria have evolved two systems to recombine extracellular DNA
www.biorxiv.org/content/10.1...
Kudos to lead authors Léo Hardy, Violette Morales and Clothilde Rousseau, and to outstanding Dalia's lab and @epcrocha.bsky.social
🧵⬇️
Massive update of preprint with @polardlab.bsky.social!
Bacteria have evolved two systems to recombine extracellular DNA
www.biorxiv.org/content/10.1...
Kudos to lead authors Léo Hardy, Violette Morales and Clothilde Rousseau, and to outstanding Dalia's lab and @epcrocha.bsky.social
🧵⬇️
How early gene expression coupled to DNA processing boosts resistance plasmid spread.
With @nfrk92.bsky.social in collab with Y. Yamaichi lab
👉 doi.org/10.64898/202...

How early gene expression coupled to DNA processing boosts resistance plasmid spread.
With @nfrk92.bsky.social in collab with Y. Yamaichi lab
👉 doi.org/10.64898/202...
We discovered that potato tubers exert intense selective pressure during Dickeya solani infection, driving the rapid emergence of mutations in the sRNA ArcZ. These mutations lead to the appearance of "social cheaters" that sweep through the pop. in as few as 30 generations.
We discovered that potato tubers exert intense selective pressure during Dickeya solani infection, driving the rapid emergence of mutations in the sRNA ArcZ. These mutations lead to the appearance of "social cheaters" that sweep through the pop. in as few as 30 generations.
www.cell.com/current-biol...

www.cell.com/current-biol...

Studying obligate predators like Bdellovibrio bacteriovorus is tricky—essential genes for predation are also essential for survival.
We expanded its genetic toolbox:
🧬promoters to fine-tune expression
🧬IPTG-inducible system
🧬CRISPRi for rapid knockdown
bit.ly/46GUn2c
1/4

Studying obligate predators like Bdellovibrio bacteriovorus is tricky—essential genes for predation are also essential for survival.
We expanded its genetic toolbox:
🧬promoters to fine-tune expression
🧬IPTG-inducible system
🧬CRISPRi for rapid knockdown
bit.ly/46GUn2c
1/4
@nfrk92.bsky.social
@narjournal.bsky.social
academic.oup.com/nar/article/...

@nfrk92.bsky.social
@narjournal.bsky.social
academic.oup.com/nar/article/...
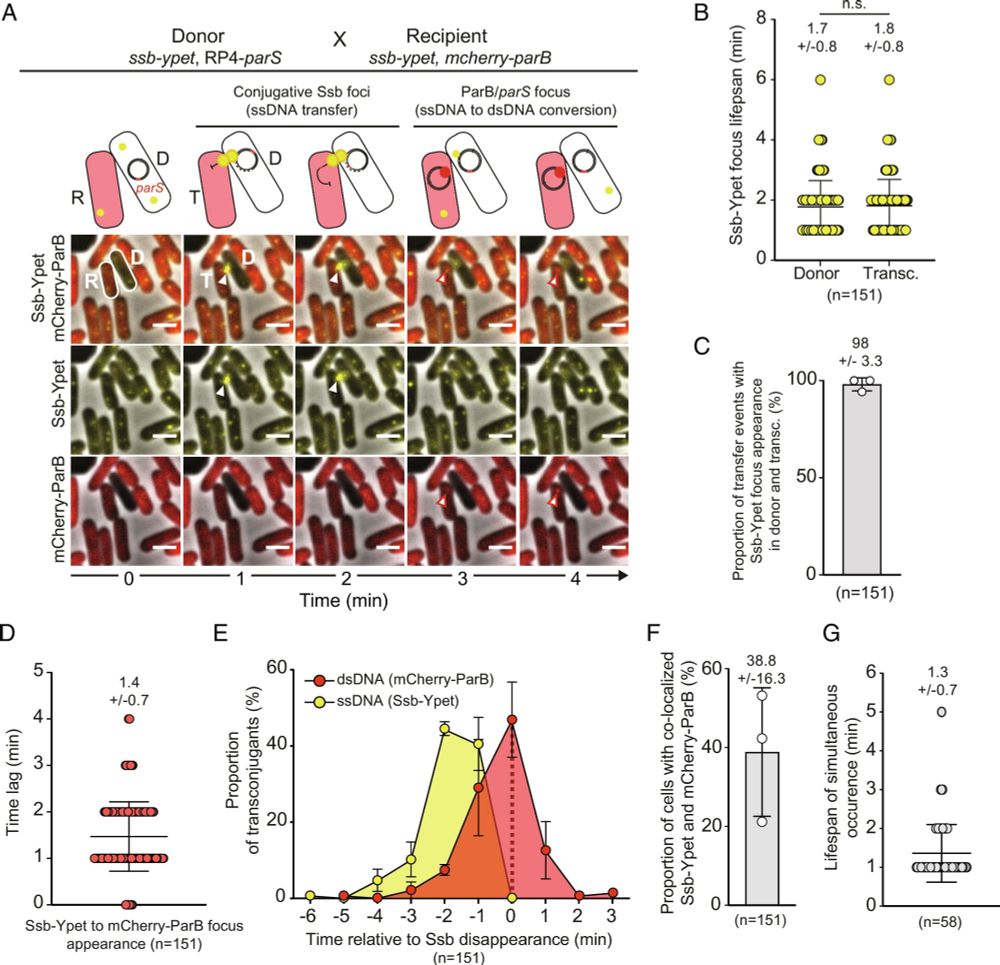
Collab with lab of @sbigot.bsky.social & Christian Lesterlin
www.pnas.org/doi/10.1073/...
Collab with lab of @sbigot.bsky.social & Christian Lesterlin
www.pnas.org/doi/10.1073/...
www.science.org/doi/10.1126/...
www.science.org/doi/10.1126/...
👉 Follow us to know about our last publications, prizes, job offers, seminars, events,...
👉 Follow us to know about our last publications, prizes, job offers, seminars, events,...




