New preprint from the lab !!! 🥳
In this work, we explore the 3D genome architecture of the virulent phage PAK_P3 during its infection cycle in P. Aeruginosa. We unveil a highly dynamic structuration as well as specific interactions patterns.
www.biorxiv.org/content/bior...
New preprint from the lab !!! 🥳
In this work, we explore the 3D genome architecture of the virulent phage PAK_P3 during its infection cycle in P. Aeruginosa. We unveil a highly dynamic structuration as well as specific interactions patterns.
www.biorxiv.org/content/bior...
✍️ Martial Marbouty
📕 @natmicrobiol.nature.com | buff.ly/1R9kqjv

✍️ Martial Marbouty
📕 @natmicrobiol.nature.com | buff.ly/1R9kqjv
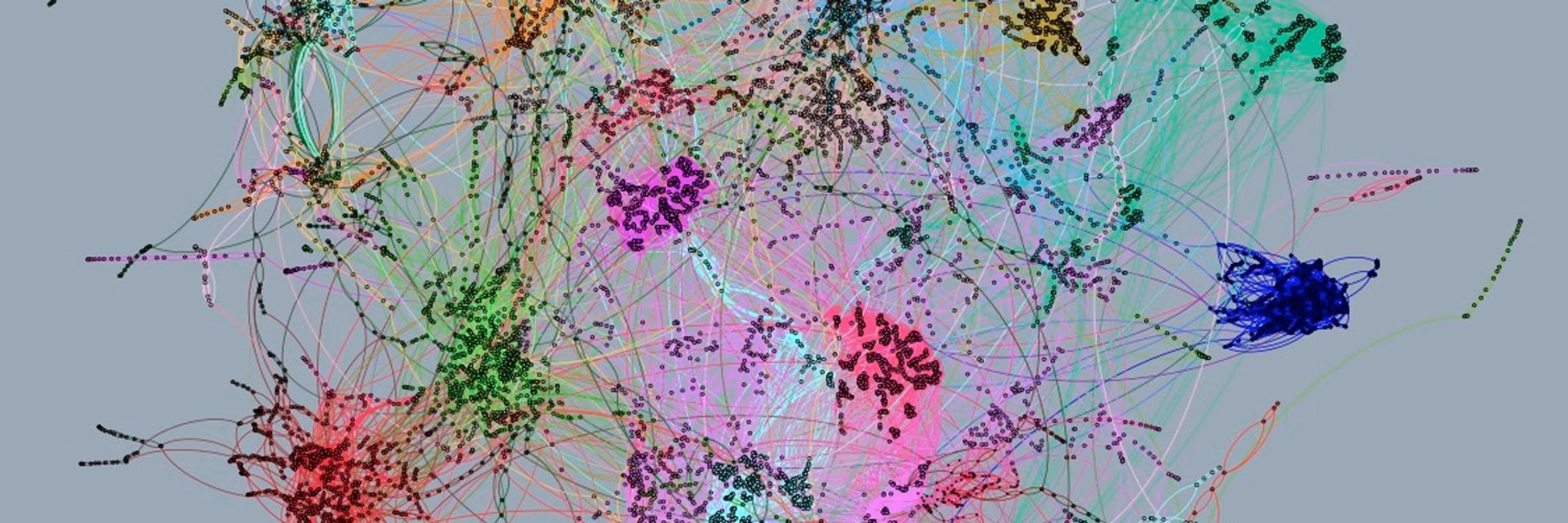
We generated and/or (re)analyzed ~100 #metagenomics Hi-C datasets to provide a comprehensive analysis of bacterial and viral metagenomes across VERY different environments.
Congratulations to all authors especially Amaury Bignaud and @mmarbout.bsky.social for the huge work!
#metaHiC
@rkoszul.bsky.social @natmicrobiol.nature.com @cnrs.fr @institutpasteur.bsky.social
www.nature.com/articles/s41...

We generated and/or (re)analyzed ~100 #metagenomics Hi-C datasets to provide a comprehensive analysis of bacterial and viral metagenomes across VERY different environments.
Congratulations to all authors especially Amaury Bignaud and @mmarbout.bsky.social for the huge work!
#metaHiC
@rkoszul.bsky.social @natmicrobiol.nature.com @cnrs.fr @institutpasteur.bsky.social
www.nature.com/articles/s41...

@rkoszul.bsky.social @natmicrobiol.nature.com @cnrs.fr @institutpasteur.bsky.social
www.nature.com/articles/s41...



